Point Cloud Optimization
import multipers as mp
from multipers.data import noisy_annulus, three_annulus
import multipers.ml.point_clouds as mmp
import multipers.ml.signed_measures as mms
import gudhi as gd
import numpy as np
import matplotlib.pyplot as plt
import torch as t
import torch
# t.autograd.set_detect_anomaly(True)
import multipers.torch.rips_density as mpt
from multipers.plots import plot_signed_measures, plot_signed_measure
from tqdm import tqdm
torch.manual_seed(1)
np.random.seed(1)
[KeOps] Warning : Cuda libraries were not detected on the system or could not be loaded ; using cpu only mode
Spatially localized optimization
The goal of this notebook is to generate cycles on the modes of a fixed measure.
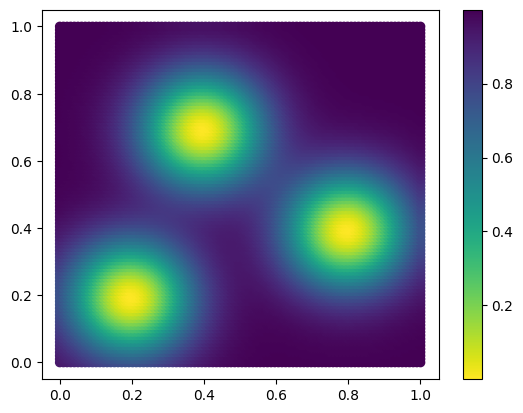
In this example, the measure is defined (in the cell below) as a sum of 3 gaussian measures.
## The density function of the measure
def custom_map(x, sigma=.17, threshold=None):
if x.ndim == 1:
x = x[None,:]
assert x.ndim ==2
basepoints = t.tensor([[0.2,0.2], [0.8, 0.4], [0.4, 0.7]]).T
out = -(t.exp( - (((x[:,:,None]- basepoints[None,:,:]) / sigma).square() ).sum(dim=1) )).sum(dim=-1)
return 1+out # 0.8 pour norme 1
x= np.linspace(0,1,100)
mesh = np.meshgrid(x,x)
coordinates = np.concatenate([stuff.flatten()[:,None] for stuff in mesh], axis=1)
coordinates = torch.from_numpy(coordinates)
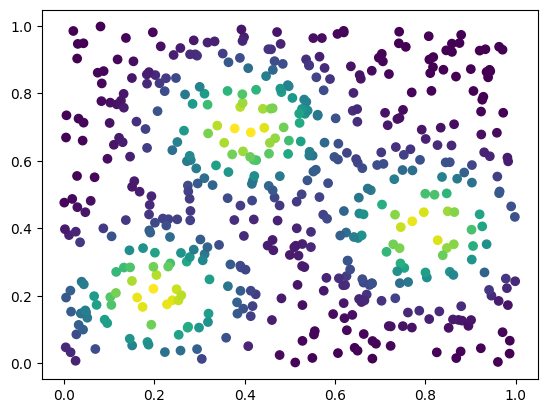
plt.scatter(*coordinates.T,c=custom_map(coordinates), cmap="viridis_r")
plt.colorbar()
<matplotlib.colorbar.Colorbar at 0x7fd9f6951160>
We start from a uniform point cloud, that we will optimize
x = np.random.uniform(size=(500,2))
x = t.tensor(x, requires_grad=True)
plt.scatter(*x.detach().numpy().T, c=custom_map(x).detach().numpy(), cmap="viridis_r")
plt.colorbar()
<matplotlib.colorbar.Colorbar at 0x7fd9f5f6ecf0>
The function_rips_signed_measure function is meant to compute signed measures from rips + function bifiltrations, in a torch-differentiable manner.
x = np.random.uniform(size=(300,2))
x = t.tensor(x, requires_grad=True)
sm_diff, = mpt.function_rips_signed_measure(
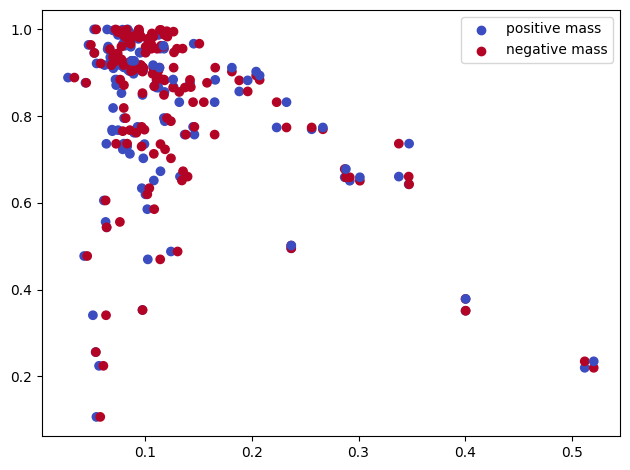
x, function=custom_map, degree=1, num_collapses=-1, plot=True, verbose=True, complex="rips");
Input is a slicer.
Reduced slicer. Retrieving measure from it...Done.
Cleaning measure...Done.
Pushing back the measure to the grid...Done.
For this example we use the following loss. Given a signed measure \(\mu\), define
where \(x := (r,d)\in \mathbb R^2\) (\(r\) for radius, and \(d\) for codensity value of the original measure) $\(\varphi(x) = \varphi(r,d) = r\times(\mathrm{threshold}-d)\)$
This can be interpreted as follows :
we maximise the radius of the negative point (maximizing the radius of cycles)
we minimize the radius of positive points (the edges of the connected points creating the cycles). This create pretty cycles
we care more about cycles that are close to the mode (the
threshold-dpart). The threshold is meant to prevent the cycles that are not close enough the the cycles to progressively stop to create loops.
threshold = .65
def softplus(x):
return torch.log(1+torch.exp(x))
# @torch.compile(dynamic=True)
def loss_function(x,sm):
pts,weights = sm
radius,density = pts.T
density = density
phi = lambda x,d : (
x
* (threshold-d)
).sum()
loss = phi(radius[weights>0], density[weights>0]) - phi(radius[weights<0], density[weights<0])
return loss
loss_function(x,sm_diff) #test that it work. It should make no error + have a gradient
tensor(0.2417, grad_fn=<SubBackward0>)
xinit = np.random.uniform(size=(500,2)) # initial dataset
x = t.tensor(xinit, requires_grad=True)
adam = t.optim.Adam([x], lr=0.01) #optimizer
losses = []
plt.scatter(*x.detach().numpy().T, c=custom_map(x, threshold=np.inf).detach().numpy(), cmap="viridis_r")
plt.show()
for i in range(101): # gradient steps
# change backend to "multipers" if you don't have mpfree
sm_diff, = mpt.function_rips_signed_measure(x, function=custom_map, degree=1, complex="weak_delaunay"); # delaunay can be changed to rips
adam.zero_grad()
loss = loss_function(x,sm_diff)
loss.backward()
adam.step()
losses.append([loss.detach().numpy()])
with torch.no_grad():
if i %10 == 1: #plot part
base=4
ncols=3
fig, (ax1, ax2, ax3) = plt.subplots(nrows=1, ncols=ncols, figsize=(ncols*base, base))
ax1.scatter(*x.detach().numpy().T, c=custom_map(x, threshold=np.inf).detach().numpy(), cmap="viridis_r", )
plot_signed_measure(sm_diff, ax=ax2)
ax3.plot(losses, label="loss")
plt.show()
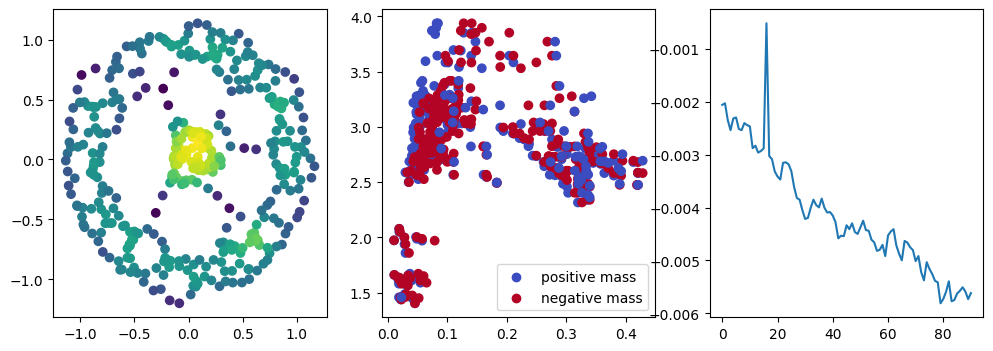
fig, (ax1, ax2, ax3) = plt.subplots(nrows=1, ncols=ncols, figsize=(ncols*base, base))
ax1.scatter(*xinit.T, c=custom_map(t.tensor(xinit)).detach().numpy(),cmap="viridis_r")
ax2.scatter(*x.detach().numpy().T, c=custom_map(x).detach().numpy(),cmap="viridis_r")
ax3.plot(losses)
plt.show()
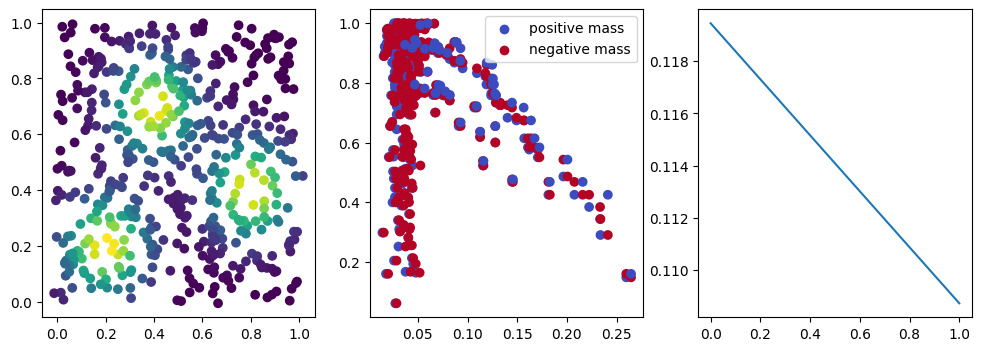
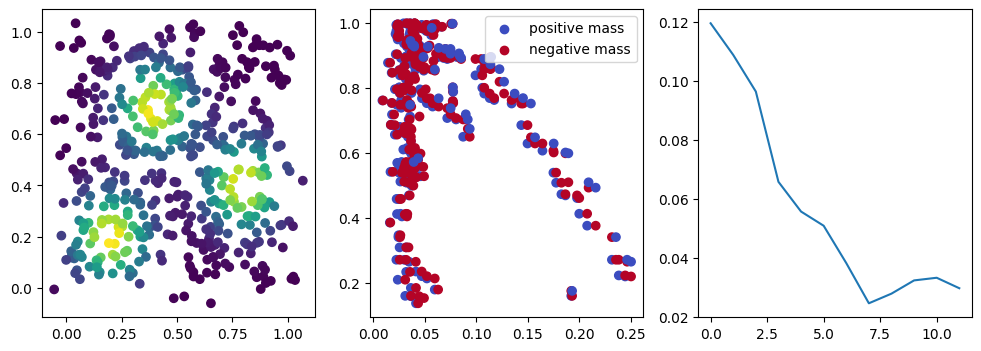
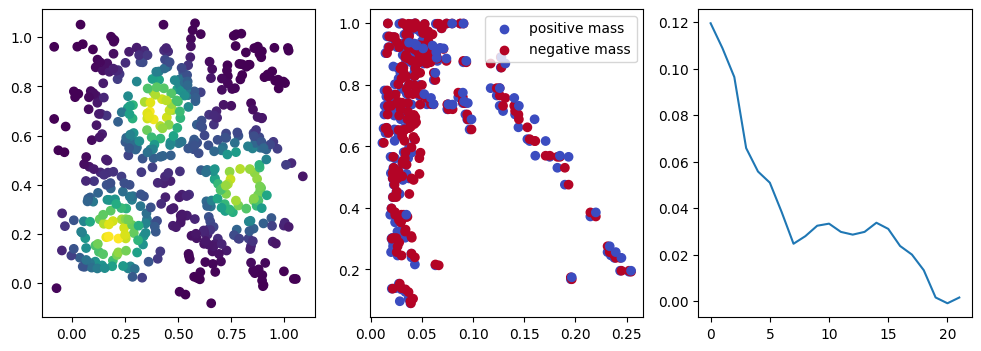
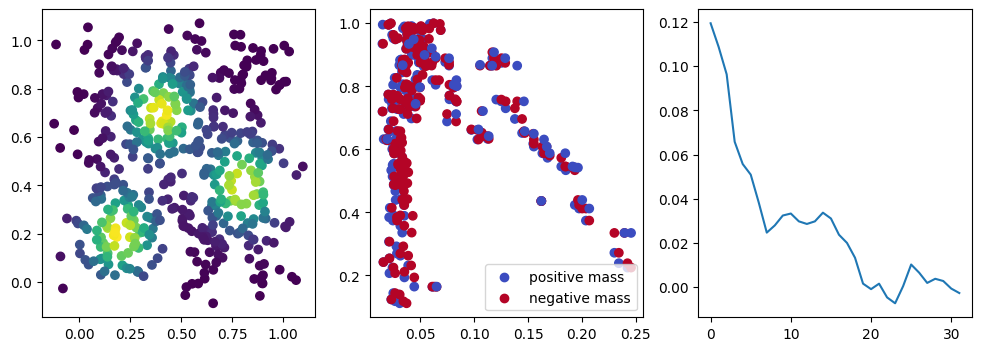
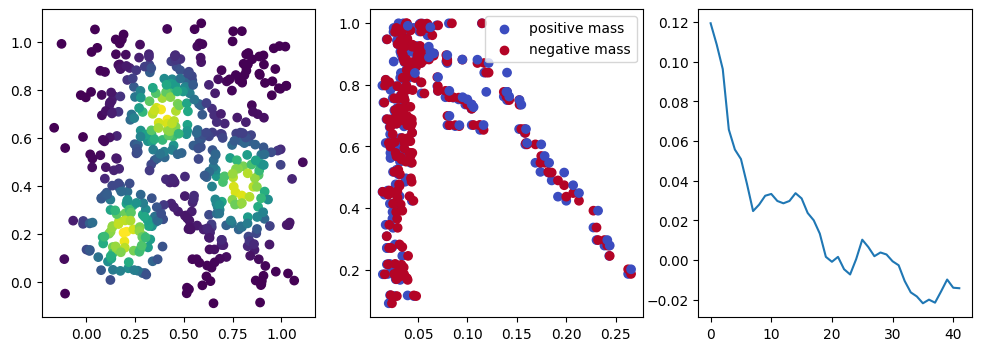
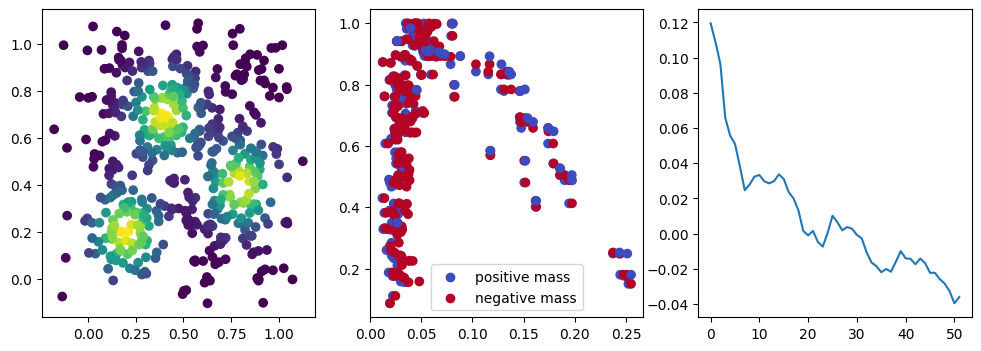
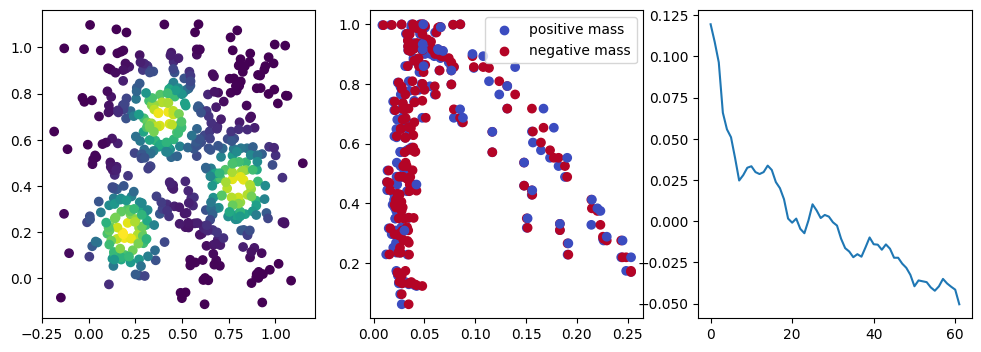
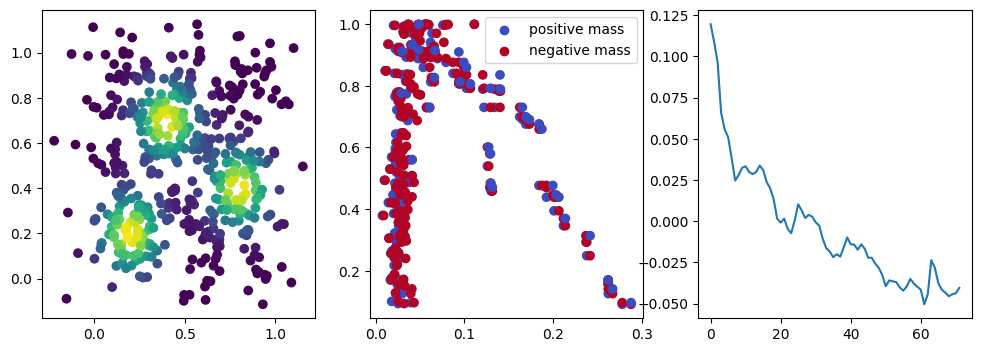
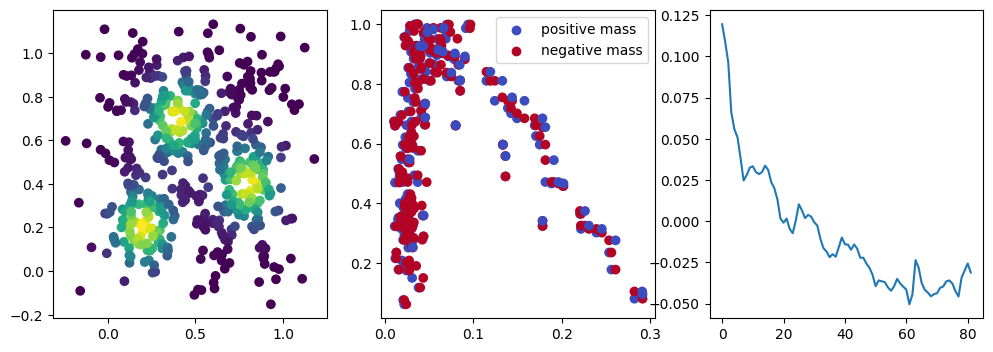
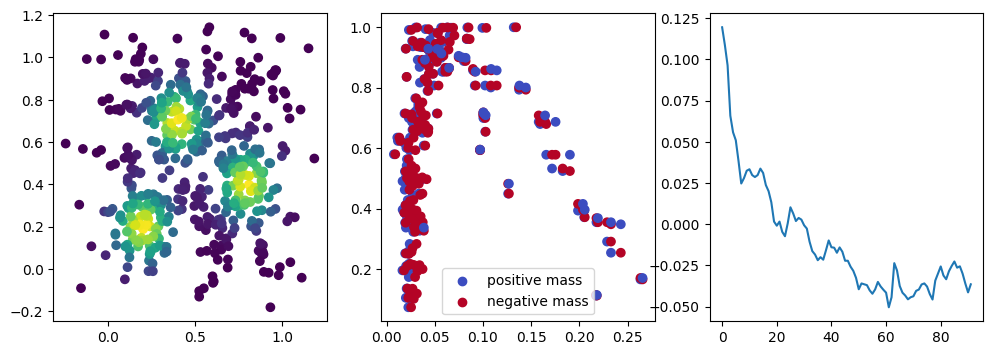
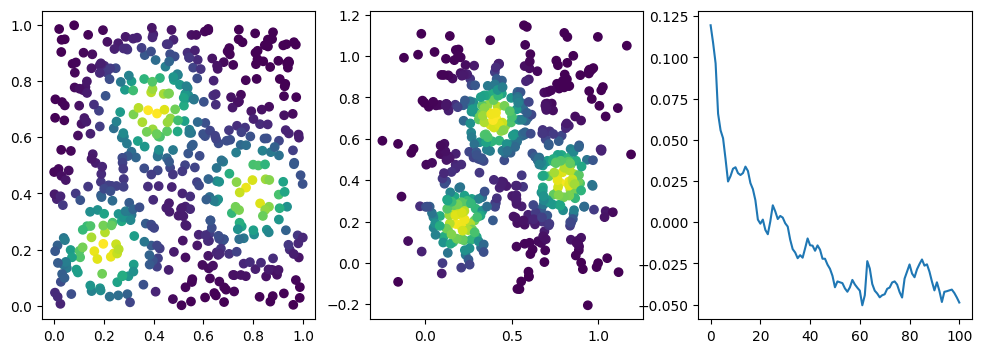
We now observe a onion-like structure around the poles of the background measure.
How to interpret this ? The density constraints (in the loss) and the radius constraints are fighting against each other, therefore, each cycle has to balance itself to a local optimal, which leads to these onion layers.
Density preserving optimization
Example taken from the paper Differentiability and Optimization of Multiparameter Persistent Homology,
and is an extension of the point cloud experiement of the optimization Gudhi notebook.
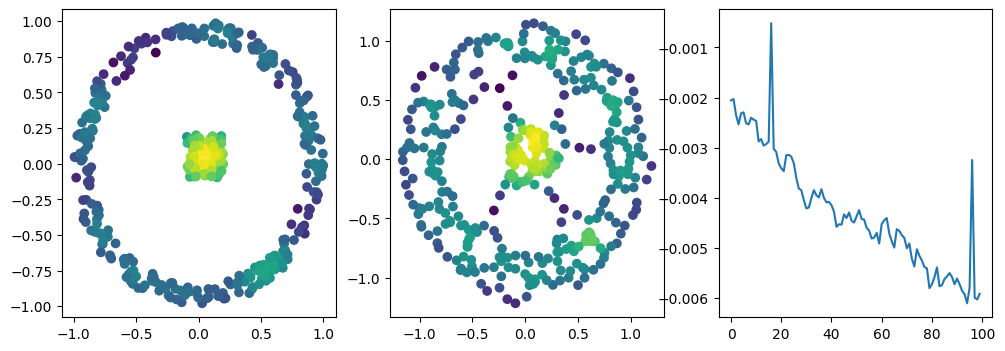
One can check from the Gudhi’s notebook that a compacity regularization term is necessary in the one parameter persistence setting; this issue will not happen when optimizing a Rips-Density bi-filtration, as we can enforce cycles to naturally balance between scale and density, and hence not diverge.
from multipers.ml.convolutions import KDE
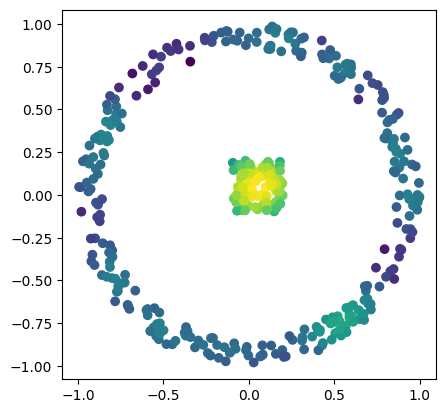
X = np.block([
[np.random.uniform(low=-0.1,high=.2,size=(100,2))],
[mp.data.noisy_annulus(300,0, 0.85,1)]
])
bandwidth = .1
custom_map2 = lambda X : -KDE(bandwidth=bandwidth, return_log=True).fit(X).score_samples(X)
codensity = custom_map2(X)
plt.scatter(*X.T, c=-codensity)
plt.gca().set_aspect(1)
def norm_loss(sm_diff,norm=1.):
pts,weights = sm_diff
loss = (torch.norm(pts[weights>0], p=norm, dim=1)).sum() - (torch.norm(pts[weights<0], p=norm, dim=1)).sum()
return loss / pts.shape[0]
x = torch.from_numpy(X).clone().requires_grad_(True)
opt = torch.optim.Adam([x], lr=.01)
losses = []
for i in range(100):
opt.zero_grad()
sm_diff, = mpt.function_rips_signed_measure(x=x, theta=bandwidth,degrees=[1], kernel="gaussian", complex="weak_delaunay")
loss = norm_loss(sm_diff)
loss.backward()
losses.append(loss)
opt.step()
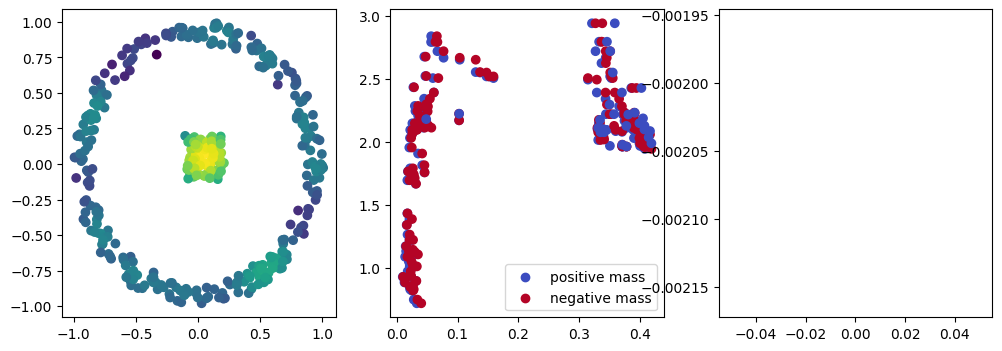
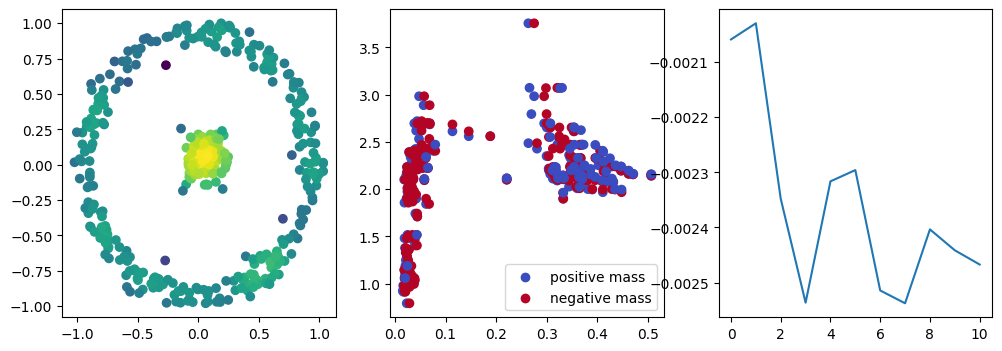
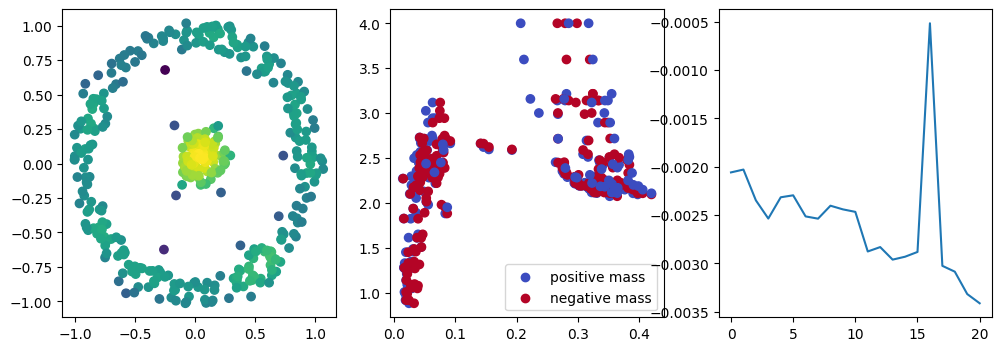
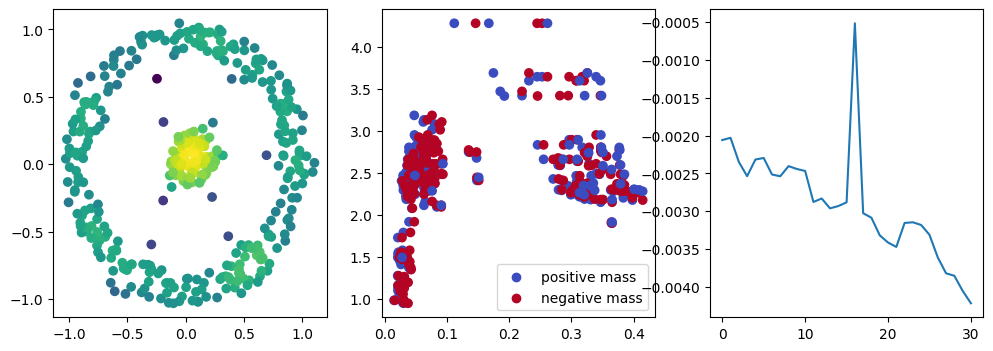
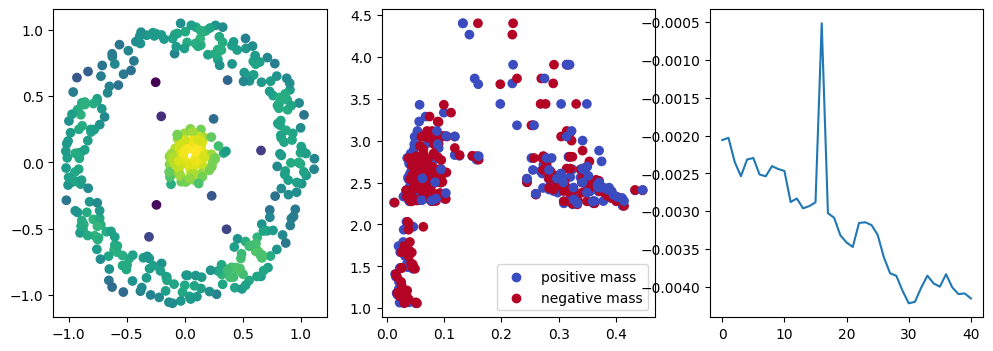
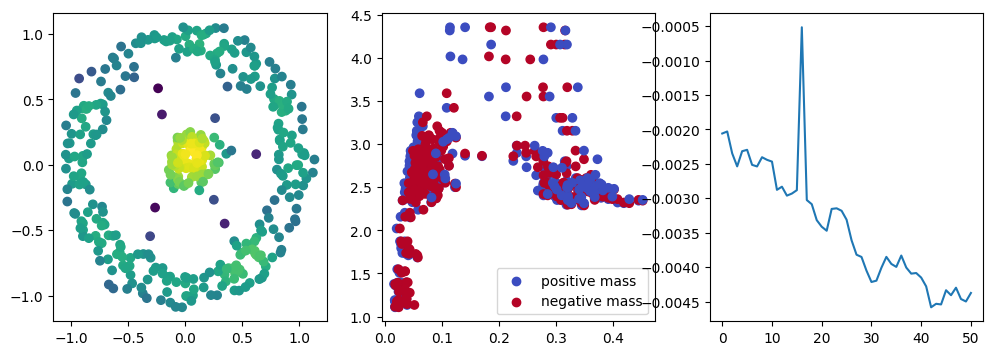
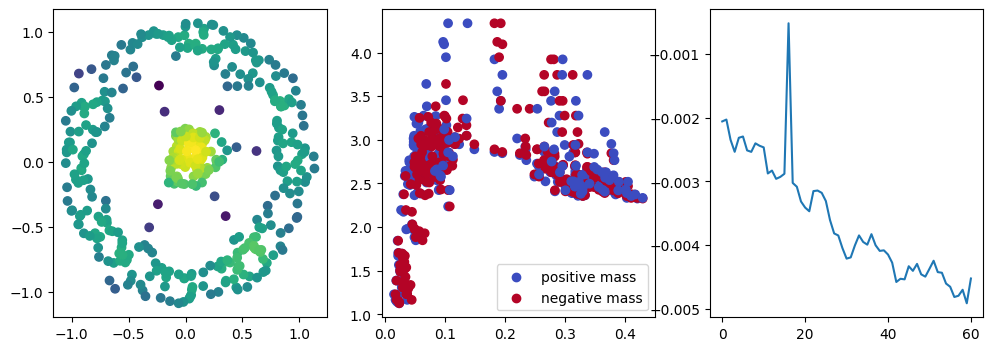
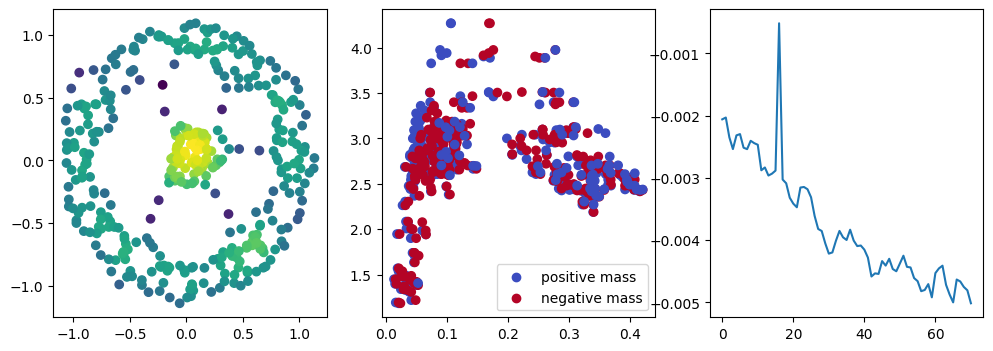
if i % 10 == 0:
with torch.no_grad():
base=4
ncols=3
fig, (ax1, ax2, ax3) = plt.subplots(nrows=1, ncols=ncols, figsize=(ncols*base, base))
ax1.scatter(*x.detach().numpy().T, c=custom_map2(x).detach().numpy(), cmap="viridis_r", )
plot_signed_measure(sm_diff, ax=ax2)
ax3.plot(losses, label="loss")
plt.show()
with torch.no_grad():
fig, (ax1, ax2, ax3) = plt.subplots(nrows=1, ncols=ncols, figsize=(ncols*base, base))
ax1.scatter(*X.T, c=custom_map2(t.tensor(X)).detach().numpy(),cmap="viridis_r")
ax2.scatter(*x.detach().numpy().T, c=custom_map2(x).detach().numpy(),cmap="viridis_r")
ax3.plot(losses)
plt.show()