Summary
During the past ten years, the biophysical modelling of the human body has been a topic of increasing interest in the field of biomedical image analysis. The aim of such modelling is to formulate personalized medicine where a digital model of an organ can be adjusted to a patient from clinical data. This virtual organ would enable to estimate the parameters which are difficult to quantify in clinical routine, such as pressure, and to test computer-based therapies that can predict the evolution of the organ over time and with therapy. Nevertheless, in order to be able to translate such an approach to clinical practice, there is a crucial demand for robust statistical methods for studying multiple cases in a patient population, in order to be able to understand the effect of different clinical factors on the anatomy and extract the significant phenomena. Such statistical analyses can both provide a predictive model and guide the biophysical approach. However, computing statistics on such complex objects (i.e. 3D shapes) is very challenging. It was traditionally relying on point based discretisation of the shapes where the point-to-point correspondence is an important limiting factor for the usability of the method. New approaches were recently developed to compute such statistics without this limitation [Durrleman et al. MICCAI 2009, MEDIA 2008, MEDIA 2009, Durrleman PhD 2009], and we present here an application of these along with an open source tool made available through the VPH Network of Excellence Toolkit that allows multiple patients to be compared and analysed using this statistical method, based on [Mansi et al, MICCAI 2009].
These tools were developed within the context of the European Health-e-Child project (2006-2009). This project regrouped 15 partners from different establishments including hospitals, research institutions, industrial institutions, and universities, and was funded by the European Commission. The aim of Health-e-Child is to develop scientific models to improve personalized child care for disease prevention, diagnosis, treatment, and therapy planning. The project allows clinicians as well as researchers to access biomedical information in order to improve paediatric care. The work we present here was motivated by the increasing demand for biomedical models integrating the information from large-scale and longitudinal data sets.
The statistical shape analysis tools are based on currents, a non-parametric representation of shapes (ere, the term ‘shape’ is used to refer to geometric data such as curves and surfaces). These tools have a wide range of applications, and provide a well-posed framework for statistical shape analysis of groups. Due to the fact that the methods do not assume point correspondences between structures (and in fact assigning landmarks to structures such as the heart are arbitrary), a wider range of data can be used. For example, one can use surfaces to model organs such as the heart, brain, and lungs, curves to model sulcal lines on the brain cortex, and sets of curves to represent fibre bundles from diffusion MRI in the brain. In brief, the basic idea is that currents probe shapes using vector fields (for further reading on the theory and mathematical details see [Durrleman PhD 2009, Durrleman et al. MEDIA 2009]). The advantage of the currents representation is that it does not assume point correspondences between structures, and as well as this, the methods can be used on any shape. Structures can then be compared at an anatomical level, since the framework is designed to measure the dissimilarity of anatomical data rather than point data. A statistical generative model is proposed where the set of shapes (curves or surfaces) are considered as random deformations of an unknown ‘ideal’ template plus some residuals. In defining shapes in this way, statistical analyses can be done on the deformations and the residuals since the framework is well-posed so that the information is controlled. Given segmented medical images, the organ anatomy of a patient is represented by a triangulated surface mesh defining the boundaries of the organ. Using these meshes, we compute an un-biased template of the population (a kind of ‘average’ shape) as well as the deformation from this template to all subjects’ surfaces. Statistical methods can then be applied to extract important phenomena. For example, Principal Component Analysis (PCA) can be applied to the transformations to obtain the principal modes of variation. Each observation of a shape can then be characterised by a shape vector built from these deformation modes. Using multiple linear regressions between the shape vectors and body surface area (BSA), the modes that are relevant to BSA are selected. Canonical Correlation Analysis (CCA) can then be applied to get a generative statistical model of the growth of the shape. The properties of the forward model allow new patients to be added to the statistical analysis.
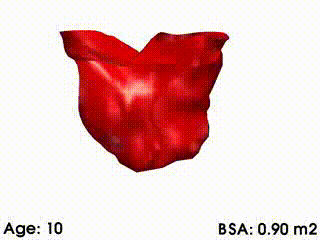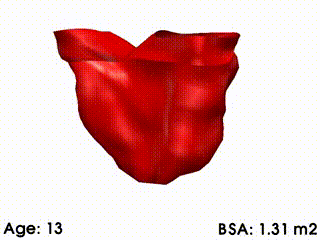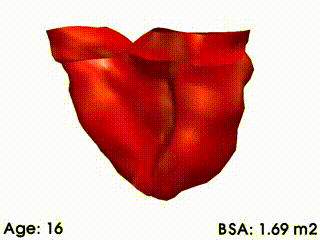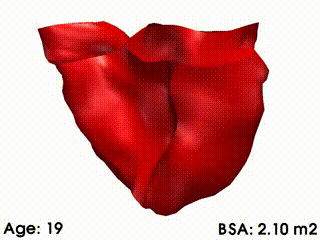
We illustrate these tools by building a template of right ventricular heart growth in patients with Tetralogy of Fallot (ToF) [Mansi et al. MICCAI 2009], a congenital heart defect that requires surgery early in infancy and affects approximately 4 out of every 10,000 babies. The long term effect of this condition is an extreme right ventricular dilation due to severe regurgitations. These patients often require repeated follow up operations. The timing of follow-up operations is crucial to ensure that the treatment is instigated before the heart becomes too deformed for intervention, while also maximizing the time between interventions because of limited life spans of the replacement valves. The clinical challenge for this condition is to understand how the heart evolves, with the main goal to be able to predict the optimal time for intervention. Our tools were used to create a model of the right ventricle growth for 18 ToF patients [Mansi et al. MICCAI 2009]. CCA was applied to quantify the amount of variation of each mode with varying body surface area (BSA is highly correlated with age for this data set). The model simulates the growth of the right ventricle as BSA increases (see movie below). While we are now in a first phase of analysis, further improvements of this model could be used to formulate a personalized prediction of heart shape, and also to predict the optimal timing of intervention. These tools could be particularly useful since they validate the predictive power of the models by comparing, retrospectively, predicted cardiac growth with that actually observed in the patient.
|
|
Mean RV remodelling observed in our population when body surface area (BSA, in m2) increases. RV dimensions globally increase while valves dilate. Simultaneously, RV free wall becomes rounder and septum more concave.
|
References
- Stanley Durrleman, Xavier Pennec, Alain Trouvé, Guido Gerig, and Nicholas Ayache. Spatiotemporal Atlas Estimation for Developmental Delay Detection in Longitudinal Datasets. In Guang-Zhong Yang, David Hawkes, Daniel Rueckert, Alison Noble, and Chris Taylor, editors, Medical Image Computing and Computer-Assisted Intervention (MICCAI'09), Part I, volume 5761 of Lecture Notes in Computer Science, London, UK, pages 297-304, September 2009. Springer. [bibtex-entry]

- Stanley Durrleman, Xavier Pennec, Alain Trouvé, and Nicholas Ayache. Sparse Approximation of Currents for Statistics on Curves and Surfaces. In Dimitris Metaxas, Leon Axel, Gábor Székely, and Gabor Fichtinger, editors, Proc. Medical Image Computing and Computer Assisted Intervention (MICCAI), Part II, volume 5242 of LNCS, New-York, USA, pages 390-398, September 2008. Springer. Note: PMID: 18982629.

- Stanley Durrleman, Xavier Pennec, Alain Trouvé, and Nicholas Ayache. Measuring Brain Variability via Sulcal Lines Registration: a Diffeomorphic Approach. In Nicholas Ayache, Sébastien Ourselin, and Anthony Maeder, editors, Proc. Medical Image Computing and Computer Assisted Intervention (MICCAI), volume 4791 of LNCS, Brisbane, Australia, pages 675-682, October 2007. Springer. Note: PMID: 18051117.

- Tommaso Mansi, Stanley Durrleman, Boris Bernhardt, Maxime Sermesant, Hervé Delingette, Ingmar Voigt, Philipp Lurz, Andrew M Taylor, Julie Blanc, Younes Boudjemline, Xavier Pennec, and Nicholas Ayache. A Statistical Model of Right Ventricle in Tetralogy of Fallot for Prediction of Remodelling and Therapy Planning. In Proc. Medical Image Computing and Computer Assisted Intervention (MICCAI'09), volume 5761 of Lecture Notes in Computer Science, London, UK, pages 214-221, September 2009. Springer.

- Stanley Durrleman, Xavier Pennec, Alain Trouvé, and Nicholas Ayache. Statistical Models on Sets of Curves and Surfaces based on Currents. Medical Image Analysis, 13(5):793-808, October 2009.

- Stanley Durrleman, Xavier Pennec, Alain Trouvé, Paul Thompson, and Nicholas Ayache. Inferring brain variability from diffeomorphic deformations of currents: an integrative approach. Medical Image Analysis, 12(5):626-637, 2008. Note: PMID: 18658005.

- S. Durrleman, Statistical models of currents for measuring the variability of anatomical curves, surfaces and their evolution, PhD thesis, Nice-Sophia Antipolis University, March 2010.
