ExoShape : shape statistics with currents
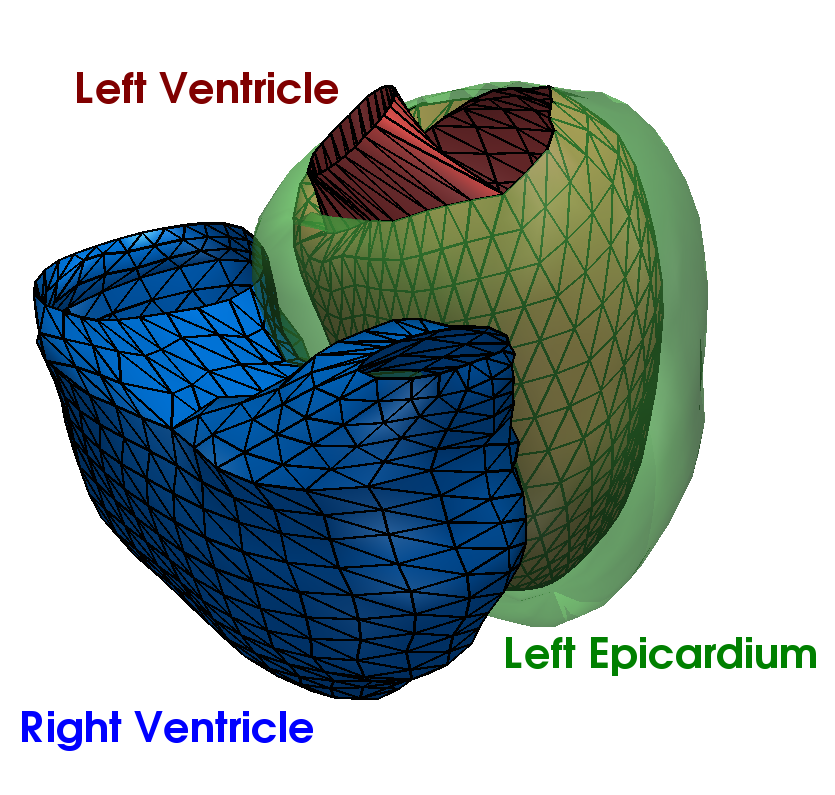
Let a population of N patients whose heart has been segmented from medical images. The cardiac anatomy of
a patient is represented by a triangulated surface mesh defining the boundaries
of the organ (Figure 1). We assume that the meshes have been rigidly aligned to
a common space [3, 4, 5]. The first
step of the analysis consists in computing an unbiased atlas of the population and
in estimating the transformations that map this atlas to the observations. We call
shapes, or observations, the computational representation of the continuous surfaces
Si and denote them by 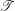 i, the superscript i standing for the patient index. It is
important to differentiate the continuous surface Si, which represents the cardiac
anatomy, and its computational representation
i, the superscript i standing for the patient index. It is
important to differentiate the continuous surface Si, which represents the cardiac
anatomy, and its computational representation 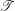 i. In the large majority of applications,
i. In the large majority of applications,
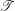 i is a triangulated mesh that approximates Si. In this framework, the
i is a triangulated mesh that approximates Si. In this framework, the 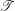 i’s are
currents.
i’s are
currents.
To create the average template of the cardiac anatomy observed in the population, we
make use of the forward strategy proposed by [1]. In the forward approach, the shapes 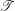 i
are modelled as the sum of an ideal template
i
are modelled as the sum of an ideal template 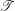 deformed by a diffeomorphic transformation
ϕi and some residuals ϵi that encode the shape features that cannot be captured by the
template. Mathematically, the forward model writes:
deformed by a diffeomorphic transformation
ϕi and some residuals ϵi that encode the shape features that cannot be captured by the
template. Mathematically, the forward model writes:
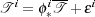 | (1) |
In this equation, * denotes the action of the transformation ϕi upon the template 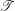 . Ideally,
. Ideally,
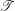 does not depend on the individuals that have been used to estimate it. In this case, the
shape information of an observation is uniquely and universally encoded by both the
transformation ϕi and the residuals ϵi. As the transformation ϕi is smooth and diffeomorphic,
it captures the shape information that is present in the template up to the transformation
smoothness. Topology changes for instance cannot be captured as they break the
diffeomorphic assumption. Similarly, shape patterns with high spatial frequency are discarded
by the smoothness of the transformation. All these “residual” features are encoded by the
residuals ϵi.
does not depend on the individuals that have been used to estimate it. In this case, the
shape information of an observation is uniquely and universally encoded by both the
transformation ϕi and the residuals ϵi. As the transformation ϕi is smooth and diffeomorphic,
it captures the shape information that is present in the template up to the transformation
smoothness. Topology changes for instance cannot be captured as they break the
diffeomorphic assumption. Similarly, shape patterns with high spatial frequency are discarded
by the smoothness of the transformation. All these “residual” features are encoded by the
residuals ϵi.
Three elements are required to apply the forward model: a consistent way to represent
shapes and residuals, a model of transformations that matches two shapes and an algorithm to
estimate a template 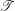 that is not biased by the population that is used to create it. In the
framework used in this work, the shape and residuals are represented by currents, an elegant
mathematical tool that defines a vector space on shapes, thus enabling statistical analyses on
shapes [2]. The transformations are modelled with the Large Deformation Diffeomorphic
Metric Mappings (LDDMM) [7]. Finally, the template
that is not biased by the population that is used to create it. In the
framework used in this work, the shape and residuals are represented by currents, an elegant
mathematical tool that defines a vector space on shapes, thus enabling statistical analyses on
shapes [2]. The transformations are modelled with the Large Deformation Diffeomorphic
Metric Mappings (LDDMM) [7]. Finally, the template 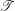 and the transformations ϕi are
estimated simultaneously with an alternate minimisation. The main concepts of the
methodology are outlined in the following paragraphs. The reader is referred to [2, 8] for
further details.
and the transformations ϕi are
estimated simultaneously with an alternate minimisation. The main concepts of the
methodology are outlined in the following paragraphs. The reader is referred to [2, 8] for
further details.
References
[1] S. Durrleman, X. Pennec, A. Trouvé, and N. Ayache, “A forward model to build unbiased atlases from curves and surfaces,” in Proc. of the International Workshop on the Mathematical Foundations of Computational Anatomy (MFCA-2008) (X. Pennec and S. Joshi, eds.), September 2008.
[2] S. Durrleman, X. Pennec, A. Trouvé, and N. Ayache, “Statistical models on sets of curves and surfaces based on currents,” Medical Image Analysis, vol. 13, pp. 793–808, Oct. 2009.
[3] F. Bookstein, “Size and shape spaces for landmark data in two dimensions,” Statistical Science, vol. 1, no. 2, pp. 181–222, 1986.
[4] I. Dryden and K. Mardia, Statistical shape analysis. Wiley New York, 1998.
[5] B. Jian and B. Vemuri, “A robust algorithm for point set registration using mixture of Gaussians,” in Proc. ICCV 2005, vol. 2, pp. 1246–1251, 2005.
[6] Y. Zheng, A. Barbu, B. Georgescu, M. Scheuering, and D. Comaniciu, “Four-chamber heart modeling and automatic segmentation for 3-d cardiac ct volumes using marginal space learning and steerable features,” IEEE transactions on medical imaging, vol. 27, no. 11, pp. 1668–1681, 2008.
[7] M. Vaillant and J. Glaunes, “Surface matching via currents,” in Proc. IPMI 2005, p. 381, Springer, 2005.
[8] S. Durrleman, Statistical models of currents for measuring the variability of anatomical curves, surfaces and their evolution. PhD thesis, Université Nice-Sophia Antipolis, March 2010.