Jerome Piovano
Research in Medical Imaging and Computer Vision
MumfordShahRegionModule Class Template Reference
RegionModule specialization used to drive the LevelSet using a scalar Mumford Shah region evolution. More...
#include <MumfordShahRegionModule.h>
Inheritance diagram for MumfordShahRegionModule:
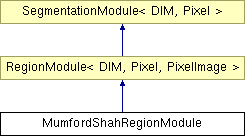
Public Types | |
|
typedef RegionModule< DIM, Pixel, PixelImage > | base |
| typedef base::LevelSet | LevelSet |
| typedef base::Index | Index |
| typedef base::Histogram | Histogram |
| typedef base::ImageType | ImageType |
| typedef base::Schemes | Schemes |
Public Member Functions | |
Constructors - Destructor | |
| MumfordShahRegionModule (ImageType *_data, float _weight=1.f, float _mu=1.f, float _dt=0.05f, int _iter=100, bool _adaptive=true) | |
| Default constructor. | |
| virtual | ~MumfordShahRegionModule () |
| Destructor. | |
Modify/Access member data | |
| float & | mu () |
| parameter of the MS equation | |
| int & | iter () |
| # of iterations of the poisson PDE | |
| float & | dt () |
| time step of the poisson PDE | |
Member functions | |
| float | value (const LevelSet &phi, const Index &ind) |
| Value of the module at a point. | |
| float | energy (const LevelSet &phi, const Index &ind) |
| Energy of the module at a point. | |
| void | init (const LevelSet &phi) |
| Initialize the module. | |
| void | update (const LevelSet &phi) |
| update the module | |
| void | save_result (const LevelSet &phi, std::string evodir, int iter) |
| save results depending on the module data | |
| void | update_mumfordshah_params (const LevelSet &phi) |
| compute the poisson PDE equations | |
Protected Member Functions | |
| float | poisson_mumford_shah_pos (const LevelSet &phi, const Index &ind) |
Compute the Mumford Shah poisson equation 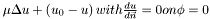 for the positive region. for the positive region. | |
| float | poisson_mumford_shah_neg (const LevelSet &phi, const Index &ind) |
Compute the Mumford Shah poisson equation 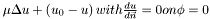 for the negative region. for the negative region. | |
Protected Attributes | |
Member data | |
| float | m_mu |
| parameter of the smoothness | |
| float | m_dt |
| time step of the gradient descent | |
| int | m_iter |
| number of iterations of the gradient descent | |
| ImageType | m_u_pos |
| smooth solution of the positive region | |
| ImageType | m_u_neg |
| smooth solution of the negative region | |
Detailed Description
template<unsigned DIM, typename Pixel = float, typename PixelImage = float>
class segmentation::MumfordShahRegionModule< DIM, Pixel, PixelImage >
RegionModule specialization used to drive the LevelSet using a scalar Mumford Shah region evolution.
- Note:
- marche + que moyen
Definition at line 24 of file MumfordShahRegionModule.h.
Constructor & Destructor Documentation
| MumfordShahRegionModule | ( | ImageType * | _data, | |
| float | _weight = 1.f, |
|||
| float | _mu = 1.f, |
|||
| float | _dt = 0.05f, |
|||
| int | _iter = 100, |
|||
| bool | _adaptive = true | |||
| ) | [inline] |
Default constructor.
- Parameters:
-
_data image to segment _weight weight of the module _mu parameter of the MS equation _dt time step of the poisson PDE _iter # of iterations of the poisson PDE _adaptive if the module is adaptive
Definition at line 109 of file MumfordShahRegionModule.h.
00110 :RegionModule<DIM, Pixel, PixelImage>(_data, _weight, _adaptive), m_mu(_mu), m_dt(_dt), m_iter(_iter), m_u_pos(_data->shape()), m_u_neg(_data->shape()) 00111 { }
Member Function Documentation
| void update_mumfordshah_params | ( | const LevelSet & | phi | ) | [inline] |
compute the poisson PDE equations
PARALLEL VERSION
SEQUENTIAL VERSION
SAVE RESULTS
Definition at line 178 of file MumfordShahRegionModule.h.
References LevelSet::extend_data(), RegionModule::m_data, MumfordShahRegionModule::m_dt, MumfordShahRegionModule::m_iter, MumfordShahRegionModule::m_u_neg, MumfordShahRegionModule::m_u_pos, MumfordShahRegionModule::poisson_mumford_shah_neg(), and MumfordShahRegionModule::poisson_mumford_shah_pos().
Referenced by MumfordShahRegionModule::init(), and MumfordShahRegionModule::update().
00179 { 00180 m_u_pos = 0.0f; 00181 m_u_neg = 0.0f; 00182 00183 BaseImage<DIM, Pixel> temp_u_pos(base::m_data->shape()); 00184 BaseImage<DIM, Pixel> temp_u_neg(base::m_data->shape()); 00185 00186 #ifdef PRINT_TIME 00187 timeval _time; 00188 Globals::initChrono(_time); 00189 #endif 00190 00192 00193 Index tmp[phi.size()]; 00194 int i = 0; 00195 for (typename LevelSet::template iterator<domain> it=phi.begin() ; it!=phi.end() ; ++it ) 00196 tmp[i++] = it.position(); 00197 00198 for (int t=0 ; t<m_iter ; t++) { 00199 #pragma omp parallel for 00200 for (int i = 0 ; i < phi.size() ; i++){ 00201 Index & ind = tmp[i]; 00202 if (phi(ind) > 0) temp_u_pos(ind) = poisson_mumford_shah_pos(phi, ind); 00203 else temp_u_neg(ind) = poisson_mumford_shah_neg(phi, ind); 00204 } 00205 #pragma omp parallel for 00206 for (int i = 0 ; i < phi.size() ; i++){ 00207 Index & ind = tmp[i]; 00208 if (phi(ind) > 0) m_u_pos(ind) += m_dt*temp_u_pos(ind); 00209 else m_u_neg(ind) += m_dt*temp_u_neg(ind); 00210 } 00211 00212 #if 0 00213 00214 for (int t=0 ; t<m_iter ; t++) { 00215 for (typename LevelSet::template iterator<domain> i=phi.begin() ; i!=phi.end() ; ++i ){ 00216 if (phi(i) > 0) temp_u_pos(i) = poisson_mumford_shah(m_u_pos, phi, i.position()); 00217 else temp_u_neg(i) = poisson_mumford_shah(m_u_neg, phi, i.position()); 00218 } 00219 00220 for (typename LevelSet::template iterator<domain> i=phi.begin() ; i!=phi.end() ; ++i ){ 00221 if (phi(i) > 0) m_u_pos(i) += m_dt*temp_u_pos(i); 00222 else m_u_neg(i) += m_dt*temp_u_neg(i); 00223 } 00224 #endif 00225 00226 #if 0 00227 ImageType tmp1(phi.shape()); 00228 ImageType tmp2(phi.shape()); 00229 for (typename LevelSet::template iterator<domain> i=phi.begin() ; i!=phi.end() ; ++i ){ 00230 if (phi(i) > 0) { tmp1(i) = temp_u_pos(i); tmp2(i) = m_u_pos(i);} 00231 else { tmp1(i) = temp_u_neg(i); tmp2(i) = m_u_neg(i);} 00232 } 00233 00234 std::ostringstream name; 00235 std::ofstream file1; 00236 std::ofstream file2; 00237 00238 name.str(""); 00239 name << "residus_" << std::setw(6) << std::setfill('0') << t << ".inr"; 00240 file1.open(name.str().c_str()); 00241 file1 << format("inrimage") << tmp1 ; 00242 file1.close(); 00243 std::cout << " - Saved " << name.str() << " ..." << std::endl; 00244 00245 name.str(""); 00246 name << "poisson_" << std::setw(6) << std::setfill('0') << t << ".inr"; 00247 file2.open(name.str().c_str()); 00248 file2 << format("inrimage") << tmp2 ; 00249 file2.close(); 00250 std::cout << " - Saved " << name.str() << " ..." << std::endl; 00251 #endif 00252 } 00253 00254 #ifdef PRINT_TIME 00255 Globals::printChrono(" - poisson PDE : ", _time); 00256 Globals::initChrono(_time); 00257 #endif 00258 00259 // extension of m_u_pos in the negative region, and m_u_neg in the positive 00260 phi.extend_data(m_u_pos,-1); 00261 phi.extend_data(m_u_neg,+1); 00262 00263 #ifdef PRINT_TIME 00264 Globals::printChrono(" - data extension in band : ", _time); 00265 #endif 00266 }
The documentation for this class was generated from the following file:
- /home/jpiovano/src/Odyssee++/trunk/Libs/LevelSet/src/SegmentationModule/MumfordShahRegionModule.h