|
CELIA3D
1.0
Fluid-structure interaction using cut-cells
|
|
CELIA3D
1.0
Fluid-structure interaction using cut-cells
|
Definition of class Grille. More...
#include <fluide.hpp>
Public Member Functions | |
| Grille () | |
| Default constructor. More... | |
| Grille (int Nx0, int Ny0, int Nz0, double dx0, double x0, double dy0, double y0, double dz0, double z0) | |
| Overload of the constructor. More... | |
| ~Grille () | |
| Destructor. More... | |
| void | affiche () |
| Test display. More... | |
| void | affiche (string r) |
| Test display. More... | |
| Cellule | cellule (int i, int j, int k) |
| Access cell (i,j, k). More... | |
| Cellule | in_cell (const Point_3 &p) |
| Access the cell containing point p. More... | |
| void | in_cell (const Point_3 &p, int &i, int &j, int &k, bool &interieur) |
| Access cell containing point p. More... | |
| void | Init () |
| Definition of initial conditions. More... | |
| double | pas_temps (double t, double T) |
| Computation of the fluid time-step. More... | |
| void | BC () |
| Boundary conditions. More... | |
| double | Masse () |
| Computation of the total fluid mass. More... | |
| double | Impulsionx () |
| Computation of the total fluid x-momentum. More... | |
| double | Impulsiony () |
| Computation of the total fluid y-momentum. More... | |
| double | Impulsionz () |
| Computation of the total fluid z-momentum. More... | |
| double | Energie () |
| Computation of the total fluid energy. More... | |
| void | Impression (int n) |
| Print out results. More... | |
| void | solve_fluidx (const double dt) |
| Solve the fluid equations in the x direction. More... | |
| void | solve_fluidy (const double dt) |
| Resolution of fluid equations in the y direction. More... | |
| void | solve_fluidz (const double dt) |
| Resolution of the fluid equations in the z direction. More... | |
| void | melange (const double dt) |
| Mixing the cells with negative density or pressure with neighbours. More... | |
| void | fnumx (const double sigma, double t) |
| Computation of the numerical x-flux. More... | |
| void | fnumy (const double sigma, double t) |
| Computation of the numerical y-flux. More... | |
| void | fnumz (const double sigma, double t) |
| Computation of the numerical z-flux. More... | |
| void | corentx (double sigma) |
| Entropy correction in the x direction. More... | |
| void | corenty (double sigma) |
| Entropy correction in the y direction. More... | |
| void | corentz (double sigma) |
| Entropy correction in the z direction. More... | |
| void | Solve (const double dt, double t, int n, Solide &S) |
| Resolution of the fluid equations. More... | |
| void | Forces_fluide (Solide &S, const double dt) |
| Computation of fluid forces (Particule.Ff) and torques (Particule.Mf) applied on the Solide. More... | |
| void | Modif_fnum (const double dt) |
| Modification of fluid fluxes and discrete balance on a cell (cut-cell method). More... | |
| void | Mixage () |
| Conservative mixing of small cut-cells. More... | |
| void | Fill_cel (Solide &S) |
| Filling of the ghost cells (alpha = 1) More... | |
| void | swap_face (const Triangles &T3d_prev, const Triangles &T3d_n, const double dt, Particule &P, double &volume_test) |
| Computation of the quantity of fluid swept by an interface triangle between t-dt and t, and computation of the boundary flux. More... | |
| void | swap_face_inexact (const Triangle_3 &Tr_prev, const Triangle_3 &Tr, const Triangles &T3d_prev, const Triangles &T3d_n, const double dt, Particule &P, double &volume_test) |
| Computation of the swept quantity for an interface triangle between times t-dt and t. Computation of the boundary flux. More... | |
| void | cells_intersection_face (int &in, int &jn, int &kn, int &in1, int &jn1, int &kn1, std::vector< Bbox > &box_cells, std::vector< Cellule > &Cells) |
| List of fluid cells intersected by an interface triangle between times t-dt and t. More... | |
| void | Swap_2d (const double dt, Solide &S) |
| Computation of the quantity of fluid swept by the solid between times t-dt and t. More... | |
| void | Swap_3d (const double dt, Solide &S) |
| Cellule | voisin_fluide (const Cellule &c, bool &target) |
| Cellule | voisin_mixt (const Cellule &c, bool &target) |
| Cellule | voisin (const Cellule &c) |
| Cellule | cible (const Cellule &c, std::vector< std::vector< int > > &tab_cible) |
| void | Mixage_cible () |
| Conservative mixing of small cut-cells. More... | |
| bool | Mixage_cible2 () |
| Conservative mixing of small cut-cells. More... | |
| void | Parois_particles (Solide &S, double dt) |
| Intersection of the fluid grid with solid. More... | |
| std::vector< Point_3 > | intersection (Triangle_3 t1, Triangle_3 t2) |
Data Fields | |
| double | x |
| Position of the origin of the fluid grid. More... | |
| double | y |
| double | z |
| double | dx |
| Spatial discretization step. More... | |
| double | dy |
| double | dz |
| vector< vector< vector< Cellule > > > | grille |
| Fluid mesh. More... | |
Definition of class Grille.
| Grille::Grille | ( | ) |
Default constructor.
Variable grille represents the fluid mesh, it is a 3D array of Cellule.
| Grille::Grille | ( | int | Nx0, |
| int | Ny0, | ||
| int | Nz0, | ||
| double | dx0, | ||
| double | x0, | ||
| double | dy0, | ||
| double | y0, | ||
| double | dz0, | ||
| double | z0 | ||
| ) |
Overload of the constructor.
| (x0,y0,z0) | Position of the origin of the fluid domain |
| (dx0,dy0,dz0) | Spatial discretization step |
| (Nx0,Ny0,Nz0) | Number of fluid cells in the x, y et z directions. |
| Grille::~Grille | ( | ) |
Destructor.
| void Grille::affiche | ( | ) |
Test display.
| void Grille::affiche | ( | string | r | ) |
Test display.
| void Grille::BC | ( | ) |
Boundary conditions.
Types of BC: 1 = reflecting; 2 = periodic; 3= outflow.
| void Grille::cells_intersection_face | ( | int & | in, |
| int & | jn, | ||
| int & | kn, | ||
| int & | in1, | ||
| int & | jn1, | ||
| int & | kn1, | ||
| std::vector< Bbox > & | box_cells, | ||
| std::vector< Cellule > & | Cells | ||
| ) |
List of fluid cells intersected by an interface triangle between times t-dt and t.
Let ![$ C_1= grille[in][jn][kn]$](../../form_36.png) the cell where the interface triangle lies at time t and
the cell where the interface triangle lies at time t and ![$ C_2= grille[in1][jn1][kn1]$](../../form_37.png) the cell where it lies at time t-dt.
the cell where it lies at time t-dt.
Due to the fluid CFL condition, 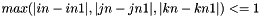 . There are therefore at most 8 cells intersected by the prism with bases the interface triangle at times t-dt (Particule.triangles_prev) and t (Particule.triangles) ):
. There are therefore at most 8 cells intersected by the prism with bases the interface triangle at times t-dt (Particule.triangles_prev) and t (Particule.triangles) ):
![$ grille[in][jn][kn]$](../../form_39.png)
![$ grille[in1][jn][kn]$](../../form_40.png)
![$ grille[in][jn1][kn]$](../../form_41.png)
![$ grille[in][jn1][kn1]$](../../form_42.png)
![$ grille[in1][jn1][kn]$](../../form_43.png)
![$ grille[in1][jn][kn1]$](../../form_44.png)
![$ grille[in][jn1][kn1]$](../../form_42.png)
![$ grille[in1][jn1][kn1]$](../../form_45.png)
| (in,jn,kn) | index of a Cellule (cell where the interface triangle lies at time t-dt (Particule.triangles_prev)) |
| (in1,jn1,kn1) | index of a Cellule (cell where the interface triangle lies at time t (Particule.triangles)) |
| box_cells | vector of Box_3 |
| Cells | vector of Cellule |
| Cellule Grille::cellule | ( | int | i, |
| int | j, | ||
| int | k | ||
| ) |
| void Grille::corentx | ( | double | sigma | ) |
Entropy correction in the x direction.
| sigma | = dt/dx : time-step/ spatial discretization step for the fluid in the x direction |
| void Grille::corenty | ( | double | sigma | ) |
Entropy correction in the y direction.
| sigma | = dt/dy : time-step/ spatial discretization step for the fluid in the y direction |
| void Grille::corentz | ( | double | sigma | ) |
Entropy correction in the z direction.
| sigma | = dt/dz : time-step/ spatial discretization step for the fluid in the z direction |
| double Grille::Energie | ( | ) |
Computation of the total fluid energy.
| void Grille::Fill_cel | ( | Solide & | S | ) |
Filling of the ghost cells (alpha = 1)
In order to compute the fluxes at the fluid-solid interface, we define a fictitious state in the cells fully occupied by solid (alpha = 1), which is taken equal to the state in the mirror cell with regards to the boundary.
Algorithm: search for the interface closest to the center of the cell (loop on all solid faces) and compute the projection of the cell center on this interface using function CGAL::projection(Point_3) .
| S | Solid |
| void Grille::fnumx | ( | const double | sigma, |
| double | t | ||
| ) |
Computation of the numerical x-flux.
| sigma | = dt/dx: time-step/fluid spatial discretization step in the x direction |
| t | current simulation time |
| void Grille::fnumy | ( | const double | sigma, |
| double | t | ||
| ) |
Computation of the numerical y-flux.
| sigma | = dt/dy: time-step/fluid spatial discretization step in the y direction |
| t | current simulation time |
| void Grille::fnumz | ( | const double | sigma, |
| double | t | ||
| ) |
Computation of the numerical z-flux.
| sigma | = dt/dz: time-step/fluid spatial discretization step in the x direction |
| t | current simulation time |
| void Grille::Forces_fluide | ( | Solide & | S, |
| const double | dt | ||
| ) |
Computation of fluid forces (Particule.Ff) and torques (Particule.Mf) applied on the Solide.
Let f an interface element, the pressure force exerted by the fluid on interface f is given by:
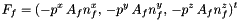
where 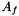 is the area of interface f,
is the area of interface f, 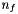 the exterior normal to f, and
the exterior normal to f, and 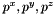 are the effective pressures in the x, y and z directions during the time-step (Cellule.pdtx, Cellule.pdty and Cellule.pdtz). The fluid torque exerted on f is given by:
are the effective pressures in the x, y and z directions during the time-step (Cellule.pdtx, Cellule.pdty and Cellule.pdtz). The fluid torque exerted on f is given by:
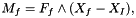
where 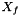 is the center of interface f and
is the center of interface f and 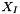 is the center of the particle containing f. These forces are transmitted to the solid as being constant during the time-step.
is the center of the particle containing f. These forces are transmitted to the solid as being constant during the time-step.
| S | Solid |
| dt | Time-step |
| void Grille::Impression | ( | int | n | ) |
Print out results.
| n | index of the output file |
| double Grille::Impulsionx | ( | ) |
Computation of the total fluid x-momentum.
| double Grille::Impulsiony | ( | ) |
Computation of the total fluid y-momentum.
| double Grille::Impulsionz | ( | ) |
Computation of the total fluid z-momentum.
| void Grille::in_cell | ( | const Point_3 & | p, |
| int & | i, | ||
| int & | j, | ||
| int & | k, | ||
| bool & | interieur | ||
| ) |
Access cell containing point p.
| p | a point |
| (i,j,k) | index of the cell containing point p |
| interieur | = true if point p is inside the fluid domain, false otherwise |
| void Grille::Init | ( | ) |
Definition of initial conditions.
| std::vector<Point_3> Grille::intersection | ( | Triangle_3 | t1, |
| Triangle_3 | t2 | ||
| ) |
| double Grille::Masse | ( | ) |
Computation of the total fluid mass.
| void Grille::melange | ( | const double | dt | ) |
Mixing the cells with negative density or pressure with neighbours.
| dt | time-step |
| void Grille::Mixage | ( | ) |
Conservative mixing of small cut-cells.
We define a small cut-cell as a cell such that 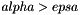 (Cellule.alpha solid occupancy ratio, and epsa: limit ratio for small cut-cells defined in parametres.hpp). In order not to modify the time-step which ensuring the CFL condition, the small cut-cells are merged with their neighbours. Denote p a small cut-cell and g a neighbouring cell such that g is totally fluid~(
(Cellule.alpha solid occupancy ratio, and epsa: limit ratio for small cut-cells defined in parametres.hpp). In order not to modify the time-step which ensuring the CFL condition, the small cut-cells are merged with their neighbours. Denote p a small cut-cell and g a neighbouring cell such that g is totally fluid~( 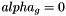 ). Define the following exchange terms:
). Define the following exchange terms:
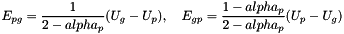
and set:

| void Grille::Mixage_cible | ( | ) |
Conservative mixing of small cut-cells.
We define a small cut-cell as a cell such that 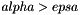 (Cellule.alpha si the solid occupancy ratio in the cell, and epsa is the maximum value accepted for CFL stability reasons and is set in parametres.hpp ). In order not to modify the time-step while keeping the original CFL condition, the small cut-cells are merged with their neighbours.
(Cellule.alpha si the solid occupancy ratio in the cell, and epsa is the maximum value accepted for CFL stability reasons and is set in parametres.hpp ). In order not to modify the time-step while keeping the original CFL condition, the small cut-cells are merged with their neighbours.
| bool Grille::Mixage_cible2 | ( | ) |
Conservative mixing of small cut-cells.
We define a small cut-cell as a cell such that 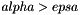 (Cellule.alpha si the solid occupancy ratio in the cell, and epsa is the maximum value accepted for CFL stability reasons and is set in parametres.hpp ). In order not to modify the time-step while keeping the original CFL condition, the small cut-cells are merged with their neighbours. This is an alternative version of Grille::Mixage_cible()
(Cellule.alpha si the solid occupancy ratio in the cell, and epsa is the maximum value accepted for CFL stability reasons and is set in parametres.hpp ). In order not to modify the time-step while keeping the original CFL condition, the small cut-cells are merged with their neighbours. This is an alternative version of Grille::Mixage_cible()
| void Grille::Modif_fnum | ( | const double | dt | ) |
Modification of fluid fluxes and discrete balance on a cell (cut-cell method).
The fluid sees the presence of the solid through this function. The final value of the state 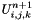 in the cell is computed using:
in the cell is computed using:
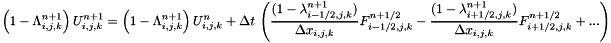
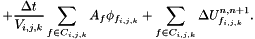
where 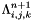 : Cellule.alpha (solid occupancy ratio in cell (i,j,k)),
: Cellule.alpha (solid occupancy ratio in cell (i,j,k)),
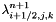 : Cellule.kappai (solid occupancy ratio on the faces of cell (i,j,k)),
: Cellule.kappai (solid occupancy ratio on the faces of cell (i,j,k)),
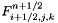 : Cellule.fluxi (flux to the right of cell (i,j,k)),
: Cellule.fluxi (flux to the right of cell (i,j,k)),
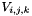 : volume of cell (i,j,k),
: volume of cell (i,j,k),
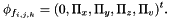
 : Cellule.phi_x, Cellule.phi_y, Cellule.phi_z, Cellule.phi_v (boundary fluxes),
: Cellule.phi_x, Cellule.phi_y, Cellule.phi_z, Cellule.phi_v (boundary fluxes),
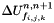 : Cellule.delta_w (swept quantity).
: Cellule.delta_w (swept quantity).
| dt | Time-step |
| void Grille::Parois_particles | ( | Solide & | S, |
| double | dt | ||
| ) |
Intersection of the fluid grid with solid.
Intersection of the fluid grid with the solid and computation of the quantities of interest: solid occupancy ratio in the cell (Cellule.alpha), solid occupancy ratio on the cell faces (Cellule.kappai, Cellule.kappaj and Cellule.kappak). Definition of the interface objects:
| S | Solide |
| dt | Time-step |
| double Grille::pas_temps | ( | double | t, |
| double | T | ||
| ) |
Computation of the fluid time-step.
| T | total simulation time |
| t | current simulation time |
| void Grille::Solve | ( | const double | dt, |
| double | t, | ||
| int | n, | ||
| Solide & | S | ||
| ) |
Resolution of the fluid equations.
Directional (Strang) splitting at each time-step.
| t | Current simulation time |
| dt | Time-step |
| n | index of the time iterations |
| void Grille::solve_fluidx | ( | const double | dt | ) |
Solve the fluid equations in the x direction.
| dt | time-step |
| void Grille::solve_fluidy | ( | const double | dt | ) |
Resolution of fluid equations in the y direction.
| dt | time-step |
| void Grille::solve_fluidz | ( | const double | dt | ) |
Resolution of the fluid equations in the z direction.
| dt | time-step |
| void Grille::Swap_2d | ( | const double | dt, |
| Solide & | S | ||
| ) |
Computation of the quantity of fluid swept by the solid between times t-dt and t.
Algorithm:
| S | Solide |
| dt | Time-step |
| void Grille::Swap_3d | ( | const double | dt, |
| Solide & | S | ||
| ) |
| void Grille::swap_face | ( | const Triangles & | T3d_prev, |
| const Triangles & | T3d_n, | ||
| const double | dt, | ||
| Particule & | P, | ||
| double & | volume_test | ||
| ) |
Computation of the quantity of fluid swept by an interface triangle between t-dt and t, and computation of the boundary flux.
Algorithm:
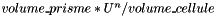 . Otherwise,
. Otherwise,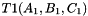 and
and 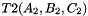 the prism bases, define points:
the prism bases, define points: 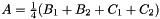 ,
, 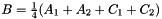 and
and 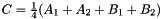 . The prism is split into the tetrahedra
. The prism is split into the tetrahedra 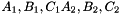 sont:
sont: 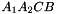 ,
, 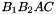 ,
, 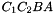 ,
, 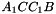 ,
, 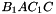 ,
, 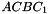 ,
, 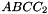 ,
, 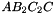 ,
, 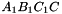 ,
, 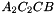 ,
, 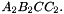
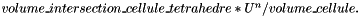
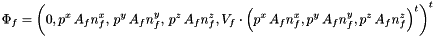
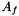 is the area of f,
is the area of f, 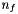 is the exterior normal to f,
is the exterior normal to f, 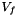 is the velocity at the center of f computed with function vitesse_parois(Point_3& ) and
is the velocity at the center of f computed with function vitesse_parois(Point_3& ) and 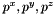 are the effective pressures in the x, y and z direction during the time-step (Cellule.pdtx, Cellule.pdty and Cellule.pdtz).
are the effective pressures in the x, y and z direction during the time-step (Cellule.pdtx, Cellule.pdty and Cellule.pdtz).| T3d_prev | Triangles_3 (interface triangles at time t: Particule.Triangles_interface) |
| T3d_n | Triangles_3 (interface triangles at time t-dt: Particule.Triangles_interface_prev) |
| dt | Time-step |
| P | Particule |
| void Grille::swap_face_inexact | ( | const Triangle_3 & | Tr_prev, |
| const Triangle_3 & | Tr, | ||
| const Triangles & | T3d_prev, | ||
| const Triangles & | T3d_n, | ||
| const double | dt, | ||
| Particule & | P, | ||
| double & | volume_test | ||
| ) |
Computation of the swept quantity for an interface triangle between times t-dt and t. Computation of the boundary flux.
Algorithm:
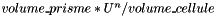 . Otherwise,
. Otherwise,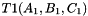 and
and 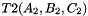 the prism bases, and define points:
the prism bases, and define points: 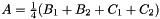 ,
, 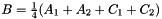 et
et 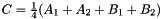 . The prism is split into the prisms
. The prism is split into the prisms 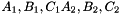 sont:
sont: 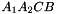 ,
, 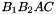 ,
, 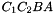 ,
, 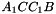 ,
, 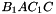 ,
, 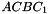 ,
, 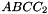 ,
, 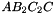 ,
, 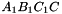 ,
, 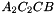 ,
, 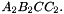
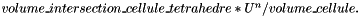
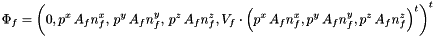
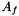 is the area of f,
is the area of f, 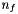 is the exterior normal to f,
is the exterior normal to f, 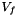 is the velocity at the center of f computed using function vitesse_parois(Point_3& ) and
is the velocity at the center of f computed using function vitesse_parois(Point_3& ) and 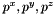 are the effective pressures in the directions x, y and z during the time-step (Cellule.pdtx, Cellule.pdty et Cellule.pdtz).
are the effective pressures in the directions x, y and z during the time-step (Cellule.pdtx, Cellule.pdty et Cellule.pdtz).| T3d_prev | Triangles_3 (interface triangles at time t: Particule.Triangles_interface) |
| T3d_n | Triangles_3 (interface triangles at time t-dt: Particule.Triangles_interface_prev) |
| dt | Time-step |
| P | Particule |
| double Grille::dx |
Spatial discretization step.
| double Grille::dy |
| double Grille::dz |
| vector< vector< vector<Cellule > > > Grille::grille |
Fluid mesh.
| double Grille::x |
Position of the origin of the fluid grid.
| double Grille::y |
| double Grille::z |
 1.8.11
1.8.11