My work aims at defining program supervision (PS) solutions adapted to the specifics of functional medical image processing (MIP) applications. To meet these requirements, I propose to integrate MIP software within the Medical Image processingAssistant (MedIA). MedIA selects, chains, tunes, runs and controls MIP steps appropriate to solve a given clinical problem, based on the expertise enclosed in its knowledge base.
My approach consists in the reuse and combination
of knowledge representations from both PS and MIP domains. On
the one hand considering PS as a problem-solving task, I have adapted the
general model of PS developed in
ORION to fit MIP
specifics. On the other hand, I have enriched the expertise enclosed in
the PS system thanks to a formalisation of
the knowledge involved in the skilled use of MIP,
both from medical and methodological points
of view. My approach is focused on the methodological aspects of
handling MIP, but also includes those pieces of knowledge involved in a
complete medical imaging examination, that impact MIP requirements
and behaviour.
The richness of data contents or the unpredictable character of MIP are taken weakly into account by previous work. Therefore, I have extended the general model of program supervision developed in previous work, thanks to a model of the role and organisation of data and processing for MIP, together with an identification of expert reasoning process and user interaction.
The use of MIP helps solving a clinical problem at a computer-level
and producing results usable at a clinical level. Knowledge about images
to analyse (i.e. acquisition information, methodological and physiological
contents), the kind of results expected from them or the underlying observed
phenomenon, altogether contribute to:
![]() transforming user's
clinical problem into a methodological objective to achieve,
transforming user's
clinical problem into a methodological objective to achieve,
![]() mapping this objective
to an appropriate and well-tuned set of MIP-level operations to execute,
mapping this objective
to an appropriate and well-tuned set of MIP-level operations to execute,
![]() presenting results
produced by MIP in a clinical way so that HCPs can compare them to their
expectations.
presenting results
produced by MIP in a clinical way so that HCPs can compare them to their
expectations.
![]() Given a clinical goal expressed in a methodological way, the usual organisation
of MIP can be abstracted as represented below.
Given a clinical goal expressed in a methodological way, the usual organisation
of MIP can be abstracted as represented below.
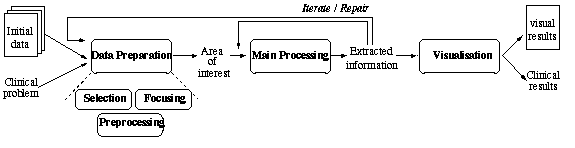
Each data transformation step involves several alternatives to consider, substeps to organise according to usual plan schemes and current conditions, MIP programs to tune well and execute, data to transmit from one program to another, results to evaluate.
![]() The overall MIP process described previously is
mainly shaped by the characteristics of the medical images to process,
which richness and role can also be modeled.
The overall MIP process described previously is
mainly shaped by the characteristics of the medical images to process,
which richness and role can also be modeled.
Used to qualify and quantify anatomical structures or functional behaviours,
medical images are multi-dimensional representations (i.e. 2 or 3 spatial
dimensions, time, energy, etc.), acquired using purpose-dependent modalities
(e.g. Computed Tomography, functional MRI, Nuclear Medicine, Ultra Sound)
and specific protocols. MIP enables to reduce the complexity of information
contained in medical images, intrinsically imprecise and incomplete, and
to improve its translation in clinical terms. Hence, MIP environment
is multi-modal, multi-dimensional and corresponds to both methodological
and medical image manipulation goals. Last, MIP successful exploitation
highly depends on the way information is presented.
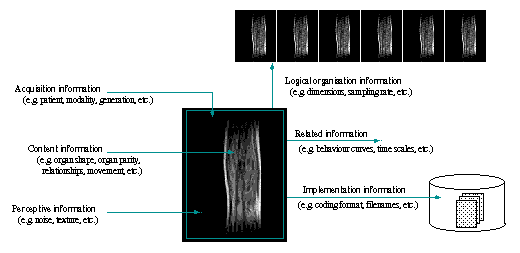
![]() MIP organisation and data features enables us to analyse experts' reasoning
process, when solving a given information extraction problem with MIP.
MIP organisation and data features enables us to analyse experts' reasoning
process, when solving a given information extraction problem with MIP.
![]() The selection of
a processing method is restricted by the nature of images to analyse and
the kind of information expected from analysis. These are related to the
clinical objective of the analysis and an a priori representation of what
images depict. The transformations that can be performed by the programs
at-hand, together with the adequacy of the observed phenomenon to their
working hypothesis are other indications to experts.
The selection of
a processing method is restricted by the nature of images to analyse and
the kind of information expected from analysis. These are related to the
clinical objective of the analysis and an a priori representation of what
images depict. The transformations that can be performed by the programs
at-hand, together with the adequacy of the observed phenomenon to their
working hypothesis are other indications to experts.
![]() First, experts sketch
the overall data transformation solution by decomposing the process into
subtasks to perform, at different levels of abstraction. As the non-exhaustive,
unpredictable character of MIP domain often impedes them to foresee the
entire process, they leave low-level decisions until solution exact instantiation.
First, experts sketch
the overall data transformation solution by decomposing the process into
subtasks to perform, at different levels of abstraction. As the non-exhaustive,
unpredictable character of MIP domain often impedes them to foresee the
entire process, they leave low-level decisions until solution exact instantiation.
![]() Then, experts take
advantage of different kinds of information and knowledge at-hand to progressively
refine decisions and run the different steps. They combine programs on
the fly, most suitably regarding actual conditions, i.e. data precise contents
and format, available programs, and the results of steps previously run.
Namely, at this stage, they are able to adapt the process to fit precise
requirements, by adding secondary processing steps (i.e. that improve data
quality, appearance or coding format, without altering data nature), or
choosing between alternatives. This reasoning is a hallmark of MIP use,
strongly determined by the presence of many sensitive parameters, and by
the visual character of result evaluation.
Then, experts take
advantage of different kinds of information and knowledge at-hand to progressively
refine decisions and run the different steps. They combine programs on
the fly, most suitably regarding actual conditions, i.e. data precise contents
and format, available programs, and the results of steps previously run.
Namely, at this stage, they are able to adapt the process to fit precise
requirements, by adding secondary processing steps (i.e. that improve data
quality, appearance or coding format, without altering data nature), or
choosing between alternatives. This reasoning is a hallmark of MIP use,
strongly determined by the presence of many sensitive parameters, and by
the visual character of result evaluation.
![]() Frequent result
evaluation and possible dynamic processing adaptation or revising in different
ways are additional key aspects of experts' way of solving problems. This
incremental reasoning process enables them to proceed with repeated trials
and errors. At each step, experts can revise previous processing or adjust
further one, depending on present results. They can decide quickly which
alternative or tuning to apply, knowing that they can easily try a better
solution if necessary. Furthermore, they can opt for simpler solutions
first, so run more costly ones only if required.
Frequent result
evaluation and possible dynamic processing adaptation or revising in different
ways are additional key aspects of experts' way of solving problems. This
incremental reasoning process enables them to proceed with repeated trials
and errors. At each step, experts can revise previous processing or adjust
further one, depending on present results. They can decide quickly which
alternative or tuning to apply, knowing that they can easily try a better
solution if necessary. Furthermore, they can opt for simpler solutions
first, so run more costly ones only if required.
![]() Experts compare
final results to the goal to achieve either by using their own experience-built
criteria, by recalling previous similar cases, or by calling upon HCPs'
specific skills.
Experts compare
final results to the goal to achieve either by using their own experience-built
criteria, by recalling previous similar cases, or by calling upon HCPs'
specific skills.
I have designed a PS system for MIP, the Medical Image processing Assistant (MedIA), suitable to take into account the rich, non-exhaustive and evolving character of the MIP domain.
![]() Main
features of the MedIA engine are exposed here (note that
a paper that describes MedIA is in preparation).
Main
features of the MedIA engine are exposed here (note that
a paper that describes MedIA is in preparation).
![]() To match experts'
incremental strategy, as in PEGASE engine,
the four PS phases of MedIA supervision
process are interleaved and the planning process is recursively
applied until primitive operators are reached, that must be executed.
To match experts'
incremental strategy, as in PEGASE engine,
the four PS phases of MedIA supervision
process are interleaved and the planning process is recursively
applied until primitive operators are reached, that must be executed.
MedIA implements a PS method that
mixes three kinds of planning strategies in a complementary way,
for the selection and organisation of processing steps to execute:
![]() abstract hierarchical
planning (reusing part of PEGASE engine) to choose and organise coarse
processing steps, based on a knowledge base of predefined hierarchical
skeletons of operators. MedIA uses initial problem specification to select
the best high-level solution skeleton for data transformation. Thanks to
expert decision criteria attached to operators, it decomposes skeletons
into a sequence of abstract steps to perform, without precisely determining
the primitive operators to use.
abstract hierarchical
planning (reusing part of PEGASE engine) to choose and organise coarse
processing steps, based on a knowledge base of predefined hierarchical
skeletons of operators. MedIA uses initial problem specification to select
the best high-level solution skeleton for data transformation. Thanks to
expert decision criteria attached to operators, it decomposes skeletons
into a sequence of abstract steps to perform, without precisely determining
the primitive operators to use.
![]() dynamic abstract
step solving, when complex operator decompositions refer to a functionality
to achieve. As for initial problem resolution, abstract steps are instanciated
thanks to selection of operators that match the current abstract problem
to solve. The functionality to achieve, the
dynamic abstract
step solving, when complex operator decompositions refer to a functionality
to achieve. As for initial problem resolution, abstract steps are instanciated
thanks to selection of operators that match the current abstract problem
to solve. The functionality to achieve, the
nature of data to process (i.e. data acquisition and contents information)
and to obtain are the criteria to filter the search space of operators
available to solve the problem.
![]() reactive operator-based
plan adaptation, to solve small run-time incompatibilities between
the state of the system (e.g. present data) and current operator requirements
(i.e. preconditions). It modifies data (i.e. perception, logical
organisation and implementation information) if
reactive operator-based
plan adaptation, to solve small run-time incompatibilities between
the state of the system (e.g. present data) and current operator requirements
(i.e. preconditions). It modifies data (i.e. perception, logical
organisation and implementation information) if
possible, e.g. by adding an intermediate step, such as preprocessing
or format conversion, selected thanks to preconditions and effects on data
description. Hence, MedIA does not require that the set of solutions be
predefined in the knowledge base.
![]() The execution
phase of MedIA tunes, runs and controls the execution of programs, using
interactive protocol-based communication with programs. It enables
their synchronised execution, on-time providing of their parameter values,
reaction to exceptional conditions, dialogue with HCPs, and visualisation
monitoring. Operator preconditions are also used for physical data existence
testing (i.e. thanks to data implementation perspective).
The execution
phase of MedIA tunes, runs and controls the execution of programs, using
interactive protocol-based communication with programs. It enables
their synchronised execution, on-time providing of their parameter values,
reaction to exceptional conditions, dialogue with HCPs, and visualisation
monitoring. Operator preconditions are also used for physical data existence
testing (i.e. thanks to data implementation perspective).
![]() MedIA evaluation
phase is complex, highly visual and interactive. MedIA automates expert-defined
criteria for methodological evaluation aspects and collaborates with the
HCP user for clinical result evaluation or labelling.
MedIA evaluation
phase is complex, highly visual and interactive. MedIA automates expert-defined
criteria for methodological evaluation aspects and collaborates with the
HCP user for clinical result evaluation or labelling.
![]() To respect the
incremental and reactive character of experts' strategy, the repair
phase is crucial. Based on the results of current step, MedIA proceeds
to subsequent refinement, iterates processing on next piece of data or
corrects the process thanks to different actions. As experts
To respect the
incremental and reactive character of experts' strategy, the repair
phase is crucial. Based on the results of current step, MedIA proceeds
to subsequent refinement, iterates processing on next piece of data or
corrects the process thanks to different actions. As experts
manipulate symbolic information until concrete steps, MedIA offers
facilities such as adjusting a parameter at a compound level, or reconsidering
the choice or the selection of an operator.
![]() Besides,
MedIA integrates the knowledge which experts rely on, within
concepts both relevant to embody the general
organisation of MIP
and the rich decisive matter enclosed in medical images,
and convenient to be exploited by the supervision
engine. In particular, data and domain objects are represented under different
perspectives, structured according
to the role of each piece of information for the supervision strategy.
Besides,
MedIA integrates the knowledge which experts rely on, within
concepts both relevant to embody the general
organisation of MIP
and the rich decisive matter enclosed in medical images,
and convenient to be exploited by the supervision
engine. In particular, data and domain objects are represented under different
perspectives, structured according
to the role of each piece of information for the supervision strategy.
![]() MedIA is currently
being implemented with the LAMA platform.
Thanks to this development environment, the realisation of MedIA engine
is straightforward. Moreover, part of the skeletal-plan refinement method
that has been developed for PEGASE takes in charge hierarchical planning
steps of MedIA algorithm, provided few modifications. I am currently working
on the dynamic aspects of MedIA algorithm, for abstract step solving and
adaptation steps. Besides, most of the knowledge concepts for expertise
representation exist and the knowledge bases
that I have developed for PEGASE are easily transposed to the additional
knowledge concepts necessary for MedIA mixed supervision strategy.
MedIA is currently
being implemented with the LAMA platform.
Thanks to this development environment, the realisation of MedIA engine
is straightforward. Moreover, part of the skeletal-plan refinement method
that has been developed for PEGASE takes in charge hierarchical planning
steps of MedIA algorithm, provided few modifications. I am currently working
on the dynamic aspects of MedIA algorithm, for abstract step solving and
adaptation steps. Besides, most of the knowledge concepts for expertise
representation exist and the knowledge bases
that I have developed for PEGASE are easily transposed to the additional
knowledge concepts necessary for MedIA mixed supervision strategy.
![]()