Both to specify needs in PS and to validate my
solutions, I study two applications, based on different approaches of MIP:
![]() factor analysis programs, to identify functional information
in temporal medical image sequences, based on their statistical properties;
factor analysis programs, to identify functional information
in temporal medical image sequences, based on their statistical properties;
![]() computer
vision programs, to perform brain segmentation, based on spatial characteristics
(e.g. anatomic) of images.
computer
vision programs, to perform brain segmentation, based on spatial characteristics
(e.g. anatomic) of images.
The construction of knowledge bases for these
applications helps better identify the nature of the knowledge involved
in the use of MIP, to design an adapted PS engine and to easy the expression
of this knowledge.
![]()
![]() Factor
analysis for functional MIP
Factor
analysis for functional MIP
In collaboration with the INSERM
Unit 494 (Quantitative medical imaging, directed by Pr.
Todd-Pokropek), Paris, France, my first study concerns the Factor Analysis
of Medical Image Sequences (FAMIS) method, used to estimate physiological
functions that underly NM/MRI dynamic sequences (see
article for SPIE
Medical Imaging'97 conference). FAMIS is a typical example of
a powerful MIP method, but complex and sparsely distributed: it solves
different clinical goals based on performing statistical data analysis
methods, involving many choices and parameters difficult to tune.
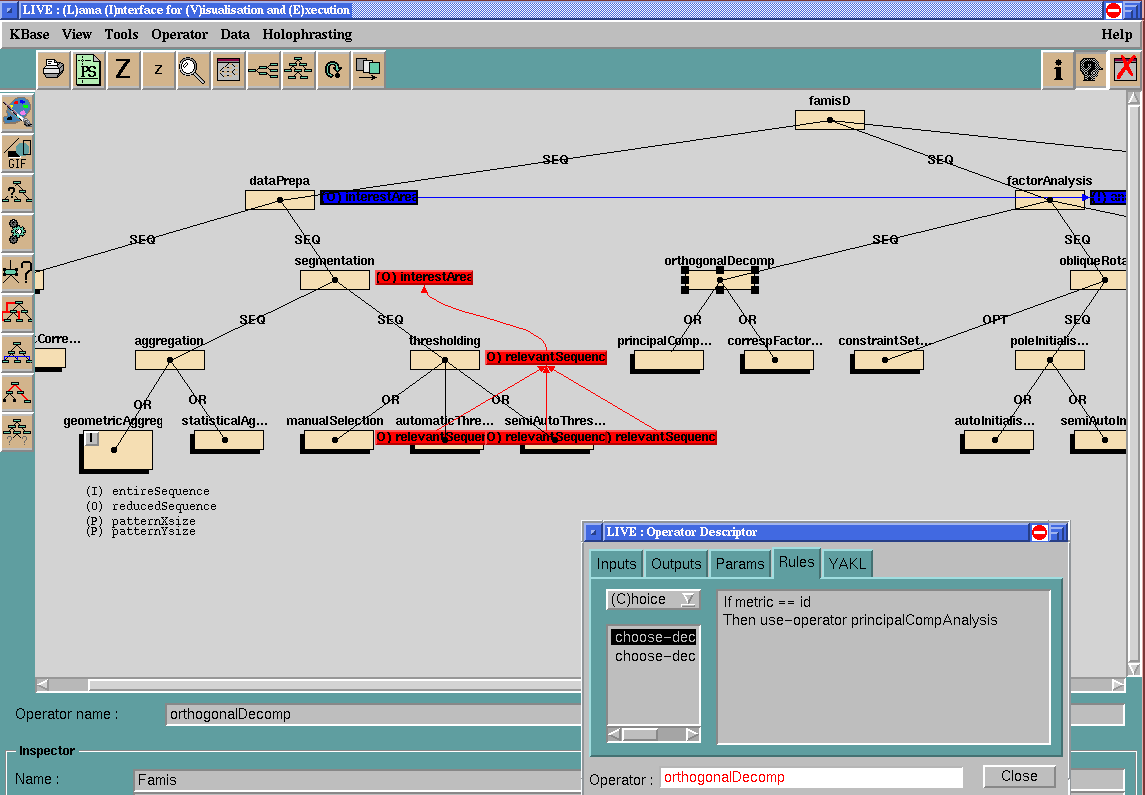
I have developed a knowledge base for FAMIS using the YAKL
language. The reference application of FAMIS in that case is osteosarcoma
chemotherapy follow-up, to predict the efficacy of the treatment.
 |
 |
 |
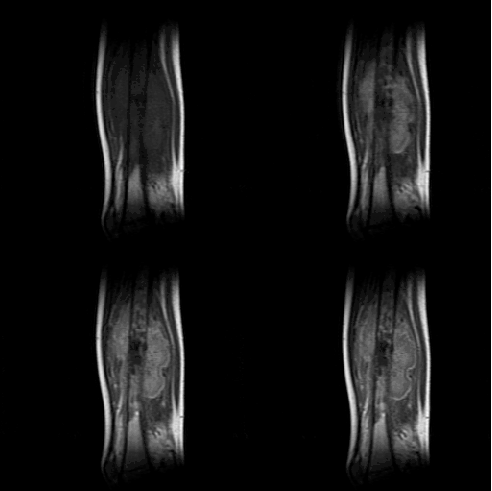
![]() FAMIS sometimes requires
the definition of one or more regions of interest (ROI) on input image
sequence: their appropriateness and characteristics are determined thanks
to expertise about the impact of focusing data on FAMIS results.
FAMIS sometimes requires
the definition of one or more regions of interest (ROI) on input image
sequence: their appropriateness and characteristics are determined thanks
to expertise about the impact of focusing data on FAMIS results.
![]() The choice of the
best way to perform FAMIS aggregation substep of data preparation is submitted
to knowledge about the way processing is influenced by the anatomical properties
of the area to analyse (e.g. organ shape, vascularisation topology).
The choice of the
best way to perform FAMIS aggregation substep of data preparation is submitted
to knowledge about the way processing is influenced by the anatomical properties
of the area to analyse (e.g. organ shape, vascularisation topology).
![]() Furthermore, if
geometric aggregation is chosen for instance, the shape and size of the
geometric pattern it uses depends on methodological information about e.g.
the quality of the image sequence, the shape and relative size of the area
of interest in the image.
Furthermore, if
geometric aggregation is chosen for instance, the shape and size of the
geometric pattern it uses depends on methodological information about e.g.
the quality of the image sequence, the shape and relative size of the area
of interest in the image.
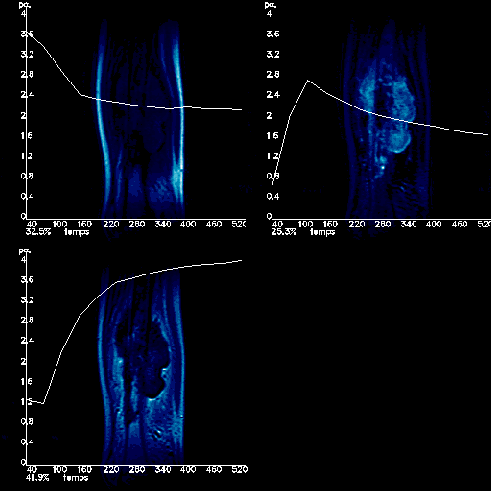
![]() When it comes to
evaluating results, expertise required is twofold: a rough idea of the
physiological activity of the analysed area and its statistical behaviour
through FAMIS, together with know-how about what can provoke FAMIS to issue
bad results (such as movement in initial image sequence).
When it comes to
evaluating results, expertise required is twofold: a rough idea of the
physiological activity of the analysed area and its statistical behaviour
through FAMIS, together with know-how about what can provoke FAMIS to issue
bad results (such as movement in initial image sequence).
![]() Whenever result
quality is not satisfying, more expertise is necessary to decide how to
correct FAMIS application.
Whenever result
quality is not satisfying, more expertise is necessary to decide how to
correct FAMIS application.
 (size: 33 Ko)
(size: 33 Ko)PEGASE engine has proven weak to take
into account the flexibility needed to handle FAMIS use. This application
is my main source of specifications and experiments for the MedIA
engine.
An example scenario of FAMIS supervision with MedIA will soon be available. ![]()
An extension of this knowledge base is in progress,
in collaboration with CERMEP-CREATIS,
within the Hospital for Neuro-cardiology, Lyon, France. In
that case, FAMIS is used to characterise myocardic perfusion on dynamic
sequences of PET or MRI images.
![]()
![]() Computer
vision for anatomic MIP
Computer
vision for anatomic MIP
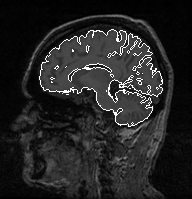
My second study concerns an application of brain anatomic segmentation on 3D MRI images based on computer vision methods (mainly mathematical morphology), in collaboration with Gregoire Malandain, from the EPIDAURE team, INRIA Sophia Antipolis. In that case, medical knowledge used by the expert is not included in programs but rather involved in expert's mental process. This study enables to define to what extent, in what form, and at what abstraction level medical knowledge is required to supervise such programs. Another interest in this application comes from expert's highly trial and error-based reasoning , as program parameter values are difficult to determine appropriately in one go. Indeed, these are very sensitive to the acquisition quality of images to process. The repair phase of PS takes here its whole importance, and extends to the necessary storage of the history of choices made for processing. Decisions also consider the global objective of the processing, e.g. to determine with which precision brain must be extracted.
 |
 |
 |
![]()