Research
Discrete
Stochastic Model
for the Generation of Axonal Trees
Abstract: In this
work we propose a 2D discrete stochastic
model for the simulation of axonal biogenesis. The model is
defined by a third order Markov Chain. The model considers
two main processes: the growth process that models the elongation
and shape of the neurites and the bifurcation process that
models the generation of branches. The growth process depends,
among other variables, on the external attraction field generated
by a chemo attractant molecule secreted by the target area. We
propose an estimation scheme of the involved parameters from
real fluorescent confocal microscopy images of single neurons
within intact adult Drosophila fly brains. Both normal neurons
and neurons in which certain genes were inactivated have been
considered (two mutations). In total, 53 images (18 normal, 21
type 1 mutant and 14 type 2 mutant) were used. The model
parameters allow us to describe pathological characteristics of
the mutated populations.
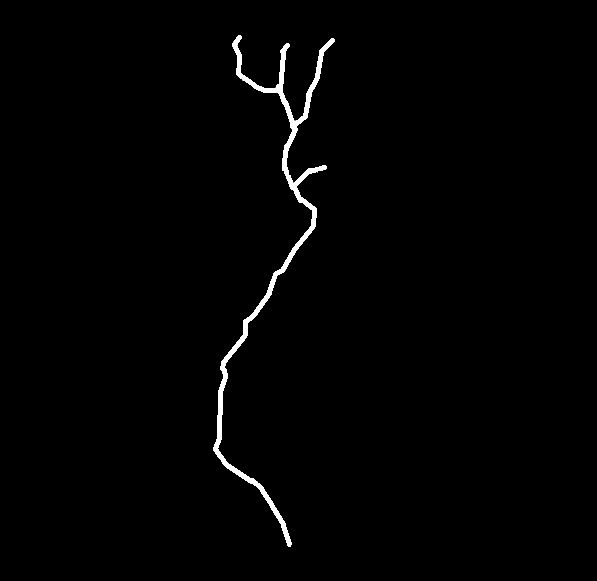
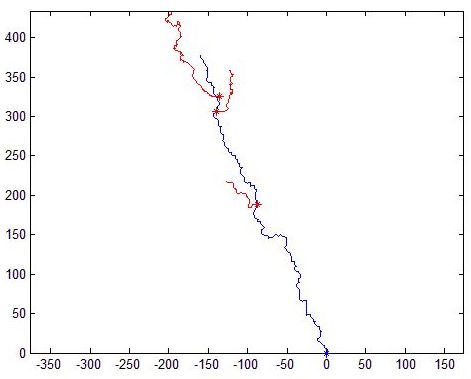
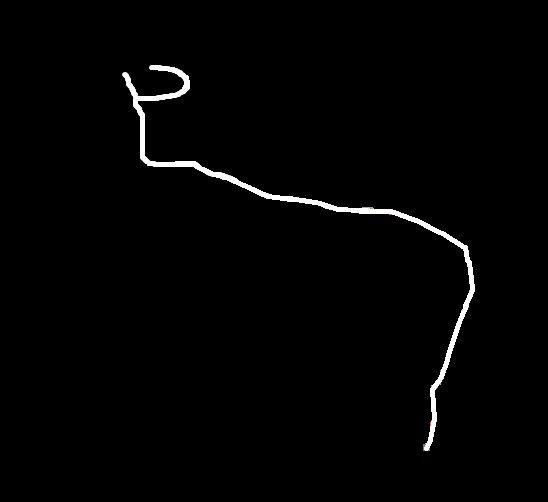
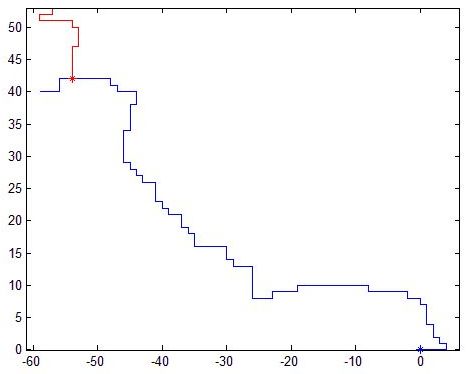
Real normal and mutant type 1 axonal trees (left top and bottom respectively) and synthetic trees (right, top and bottom) generated using the estimated parameters
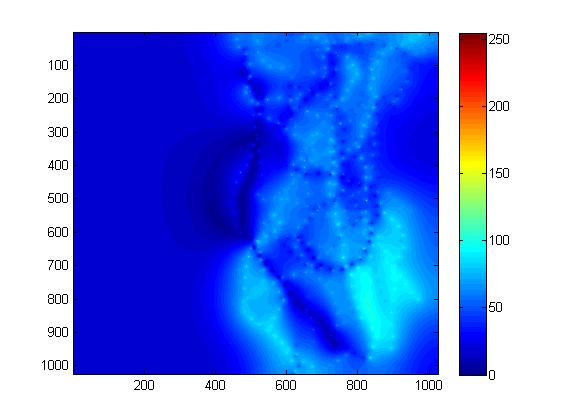
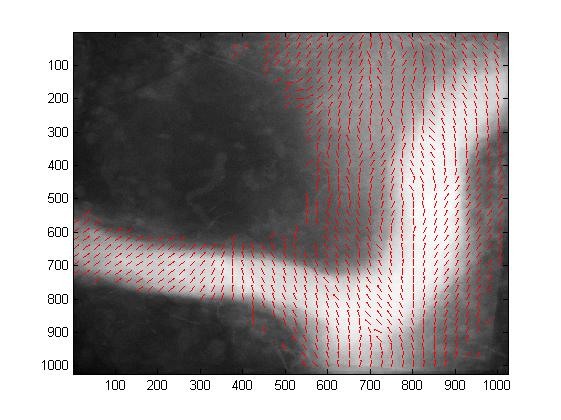
Norm (left) and direction (right) of the estimated attraction field for the normal population.
Axonal Tree Classification Using an Elastic Shape Analysis Based Distance
Abstract:
The analysis of the morphological differences between normal and
pathological neuronal structures is of paramount importance. Some
methods for the comparison of axonal trees only take into account
topological information (such as TED), while others also include
geometrical information (such as Path2Path). In a previous work, we have
presented a new method for comparing tree-like shapes based on the
Elastic Shape Analysis Framework (ESA). In this paper, we extend this
method by computing the mean shape of a population. Moreover, we propose
to evaluate and compare these 3 approaches (TED, Path2Path and ESA) with
a classification scheme based on feature computation and K-means.
We evaluate these approaches on a database of 44 real 3D
confocal microscopy images of two populations of neurons. Results show
that the proposed method distinguishes better between the two
populations.
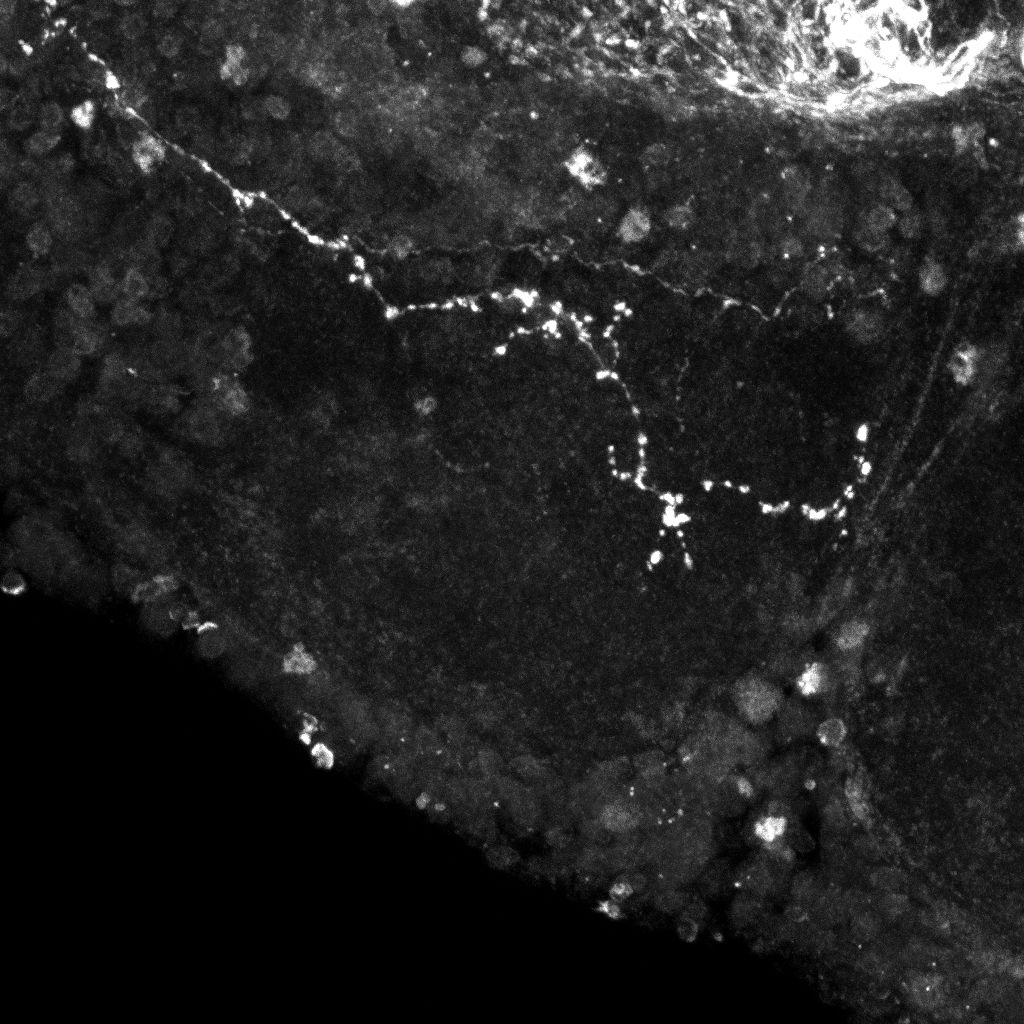
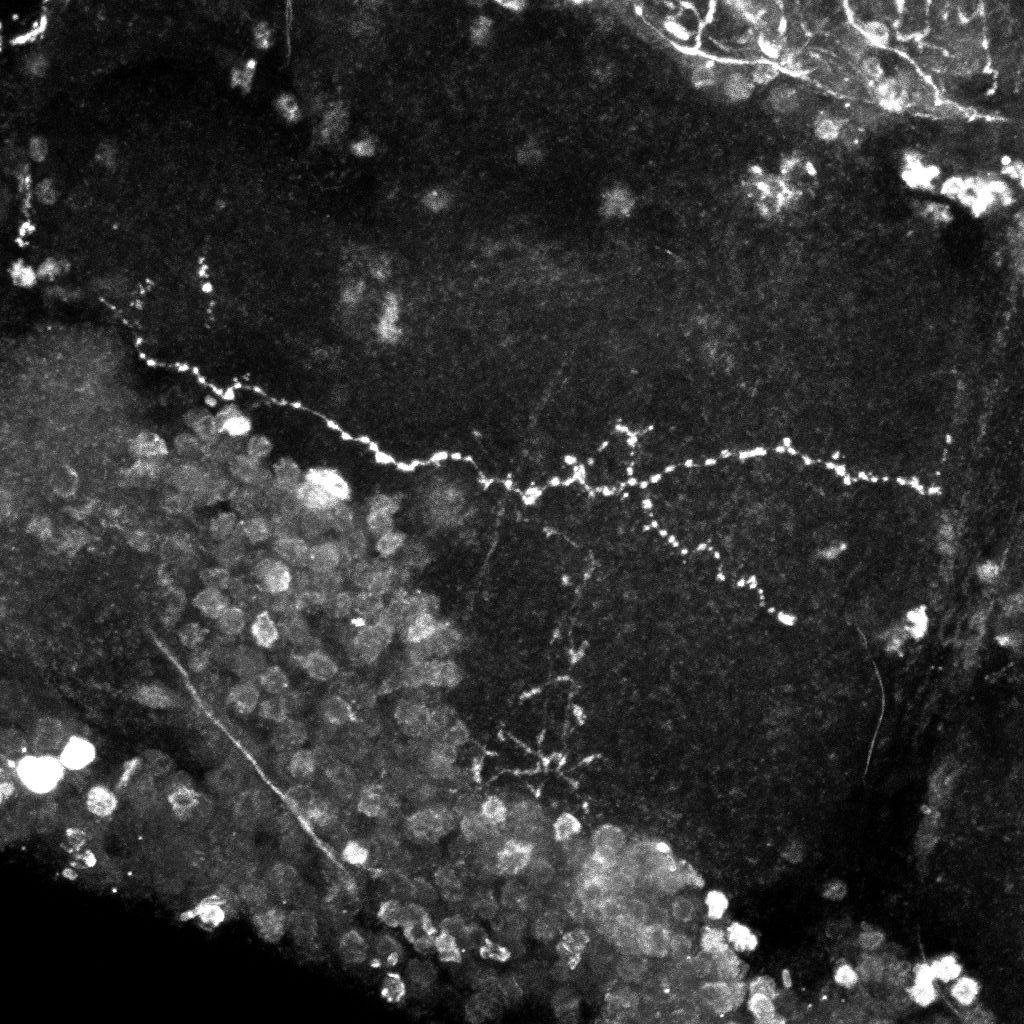
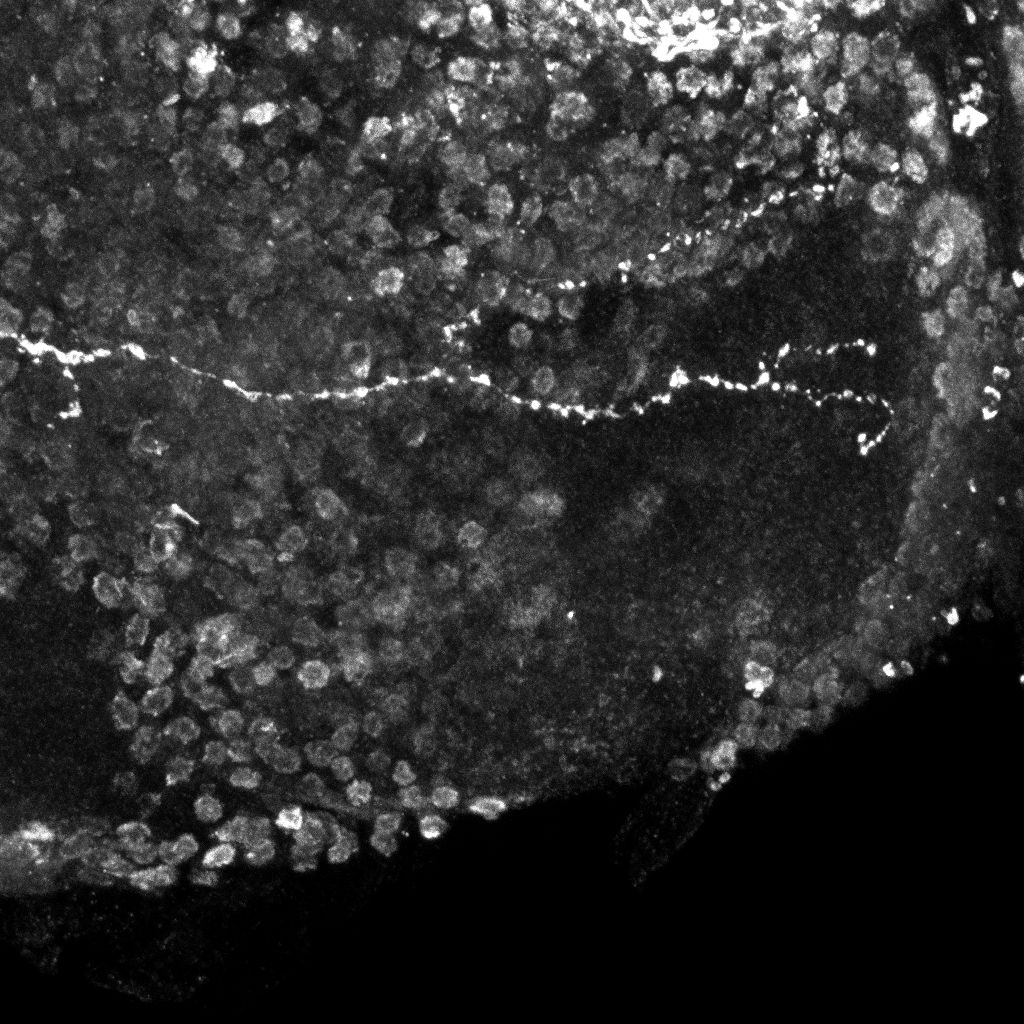
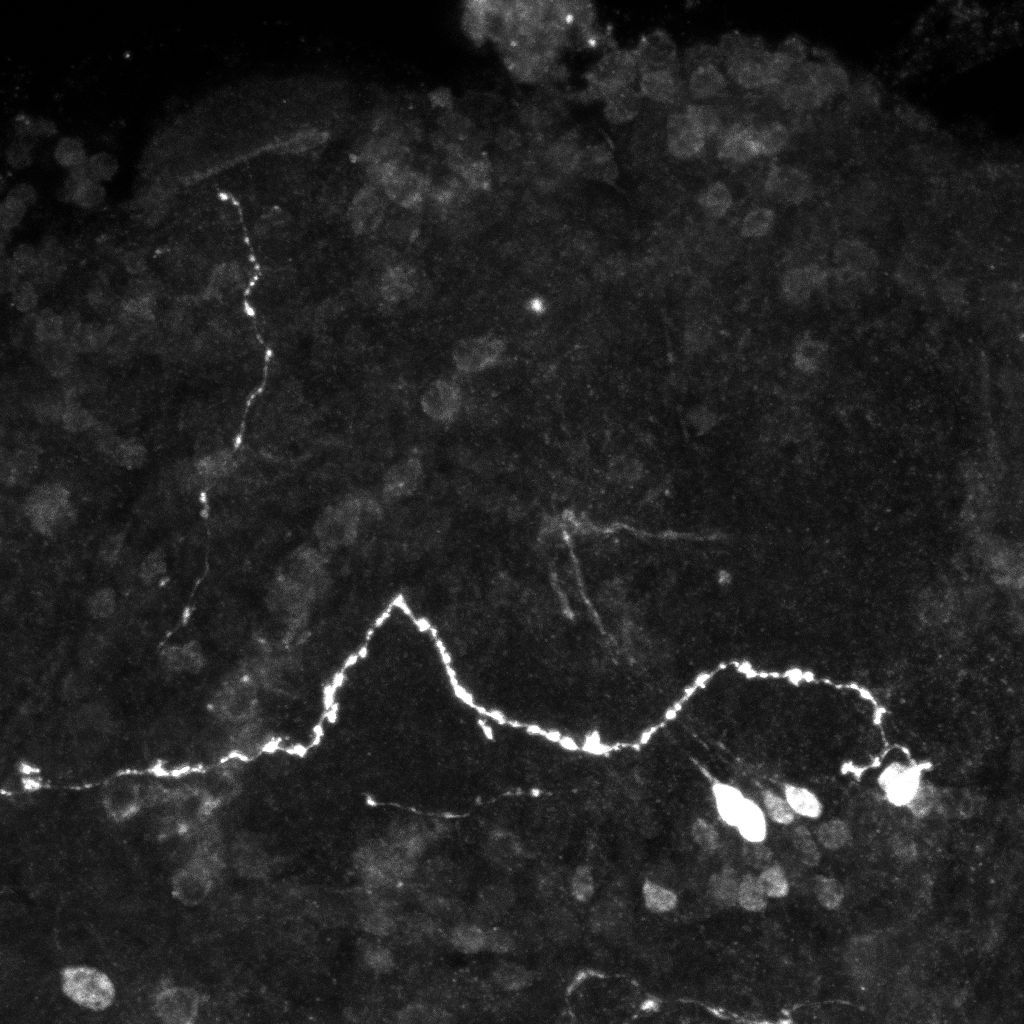
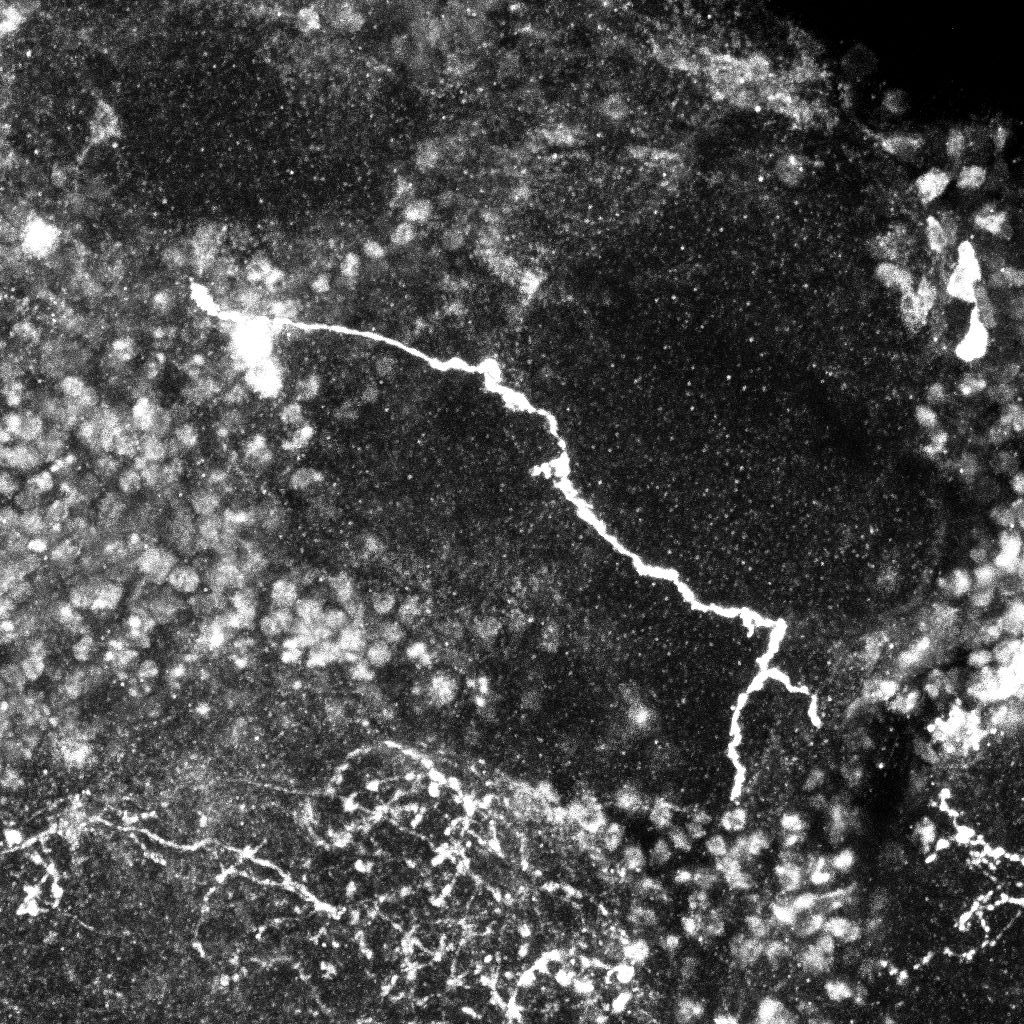

Examples of axonal trees from the normal (top) and mutant (bottom)populations (2D projections).
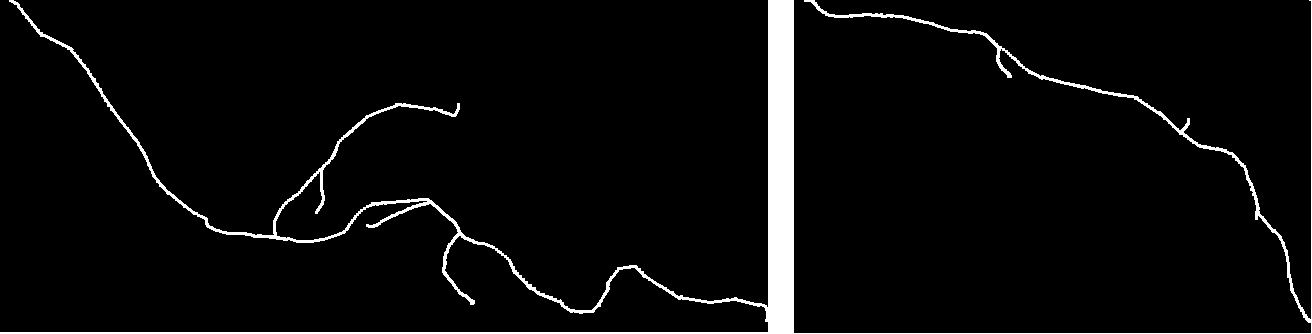
Mean normal (left) and mutant (right) axonal trees (2D projections).
Tree-like Shapes Distance Using the Elastic Shape Analysis Framework
Abstract:
The analysis and comparison of tree-like shapes is of great
importance since many structures in nature can be described by them. In
the field of biomedical imaging, trees have been used to describe
structures such as neurons, blood vessels and lung airways.Since it is
known that their morphology provides information on their functioning
and allows the characterization of pathological states, it is of
paramount importance to develop methods to analyze their shape and to
quantify differences in structures.In this paper, we present a new method
for comparing tree-like shapes that takes into account both topological
and geometrical information. It is based on the Elastic Shape
Analysis Framework, a framework originally designed for comparing shapes
of 3D closed curves in Euclidean spaces. As a first application, we used
our method for the comparison of axon morphology. The performance
was tested on a group of 44 (20 normal and 24 mutant) 3D images,
each containing one axonal tree. We have calculated inter and intra
class distances between them and implemented a basic classification
scheme.Results showed that the proposed method better distinguishes
between the two populations than a pure topological metric. Furthermore,
mean shapes can be obtained with this method.
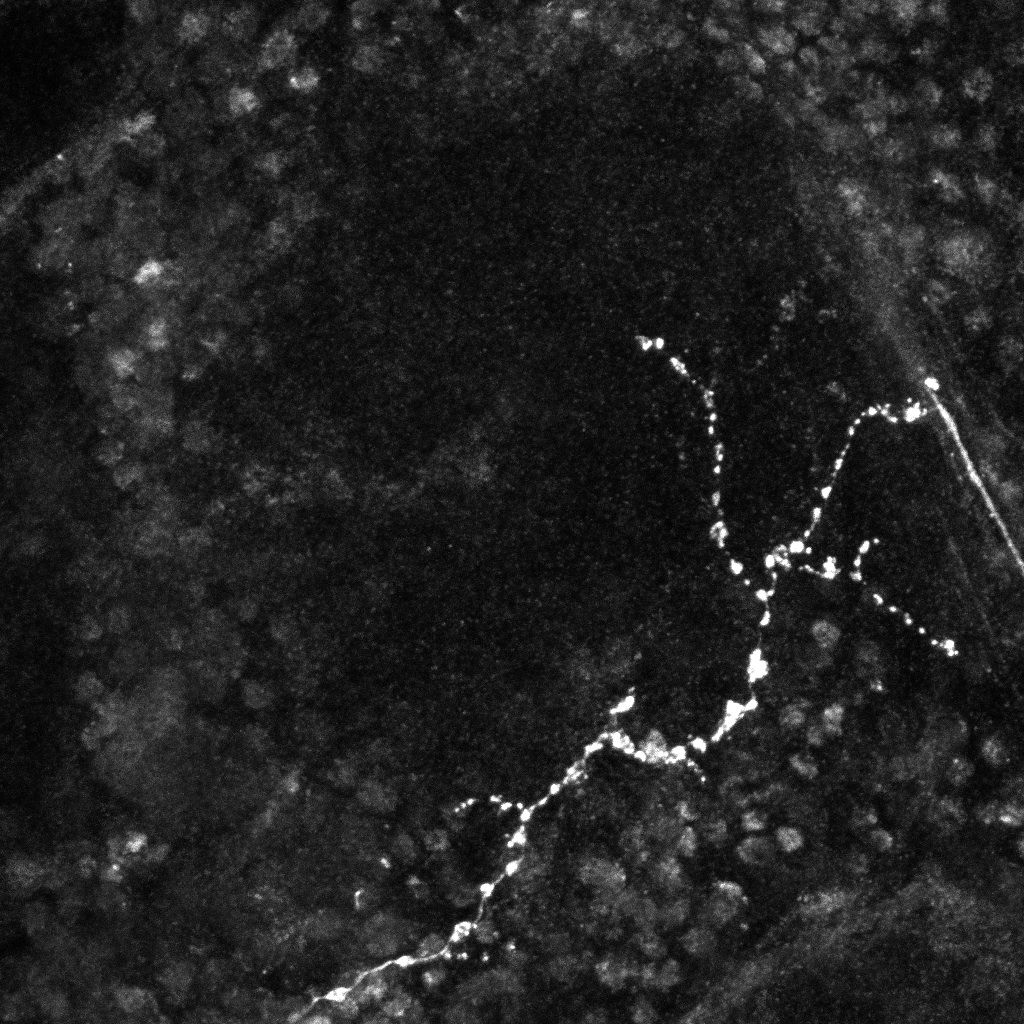
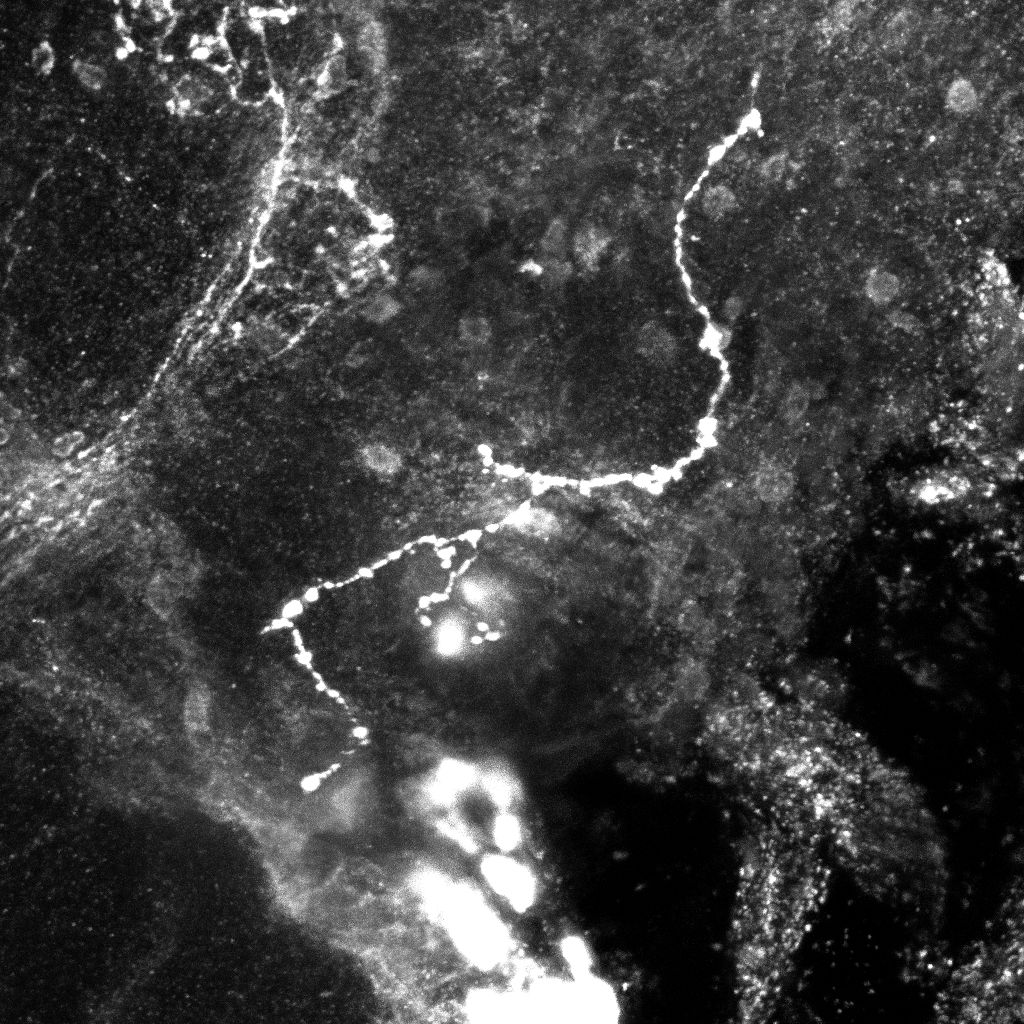

Original images (left, middle) and transformation between the two trees(2D projections).
Axon Extraction from Fluorescent Confocal Microscopy Images
Abstract:
The morphological analysis of axonal trees is an important problem in
neuroscience. The first step for such an analysis is the extraction of
the axon. Due to the high volume of generated image data and the
tortuous nature of the axons,manual processing is not feasible.
Therefore, it is necessary to develop techniques for the automatic
extraction of the neuronal structures. In this paper we present a new
approach for the automatic extraction of axons from fluorescent confocal
microscopy images. It combines algorithms for filament enhancement,
binarization,skeletonization and gap filling in a pipeline capable of
extracting the axons. The performance of the proposed method was
evaluated on real images. Results support the potential use of this
technique in helping biologists perform automatic extraction of axons
from fluorescent confocal microscopy images.

Comparison between original image (left), our result (middle) and ground truth (right) for two images (maximum intensity projections).


Original maximum intensity projection image of a normal axon (left) and extracted axon in 3D (right).
Detection and Tracking of Axonal Tips from Bi-Photon Microscopy Images
Abstract:Live cell
two-photon microscopy is an effective tool for the analysis of dynamical
processes occurring in living samples. This technique,when combined
with fuorescence, allows the detection of objects of interest in 3D
space and time. Due to the high volume of data,the automatic
analysis of the images is desired. To this end, the Marked Point Process
(MPP) detection framework was selected. MPP is a probabilistic framework
which has been successfully applied to the detection of objects in
different image processing applications. Its main advantages are that
the number of objects to be detected can be unknown, and that geometric
constraints on the objects can be easily modeled. As a first
approximation, we proposed a 3D MPP model of spheres to extract the
axonal extremities. We define a prior energy designed to penalizes (but
not forbid) overlaps, and a data energy based on the Bhattacharya
distance between the distributions. Once the energy function has been
defined, the issue is to find the configuration U(X)that minimizes it.
Due to the intricate nature of U(x), usual minimization algorithms
cannot be applied. Therefore, we have chosen Multiple Births and Deaths
(MBD). The method was evaluated on a set of real 3D+t images. Although
results show there is still work to be than in this area, they suggest
this approach could be appropriate for solving the extraction problem.
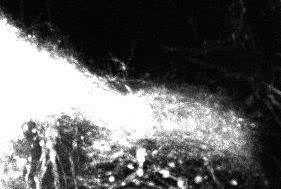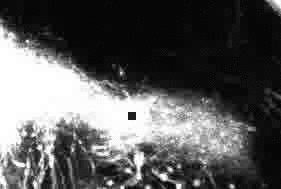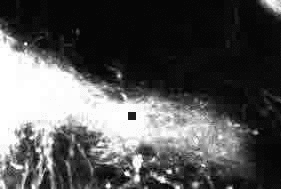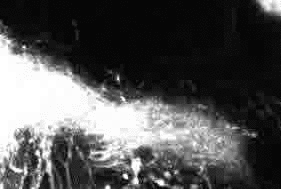
Dynamic 3D+time image sequences of developing neurons.
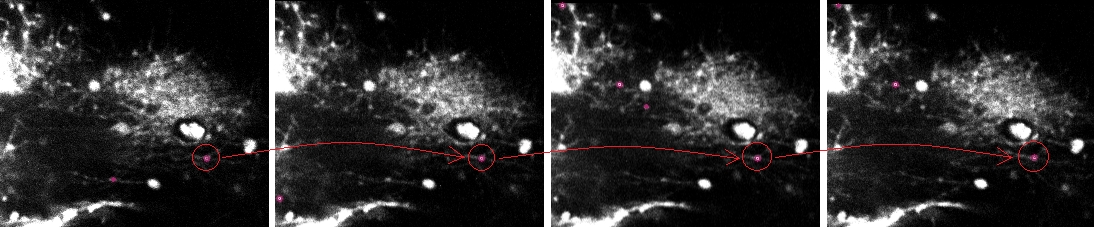
Tracking result for four consecutive frames (left to right).

Dynamic 3D+time image sequences of developing neurons.

Tracking result for four consecutive frames (left to right).
Integrated Software for the Detection of Epileptogenic Zones in Refractory Epilepsy
Abstract: We
present an integrated software designed to help nuclear
medicine physicians in the detection of epileptogenic zones (EZ) by
means of ictal-interictal SPECT and MR images. This tool was designed to
be flexible, friendly and efficient. A novel detection method was
included(A-contrario) along with the classical detection method
(Subtraction analysis). The software’s performance was evaluated with
two separate sets of validation studies: visual interpretation of
12patient images by an experimented observer and objective analysis
of virtual brain phantom experiments by proposed numerical observers.
Our results support the potential use of the proposed software to
help nuclear medicine physicians in the detection of EZ in clinical
practice.
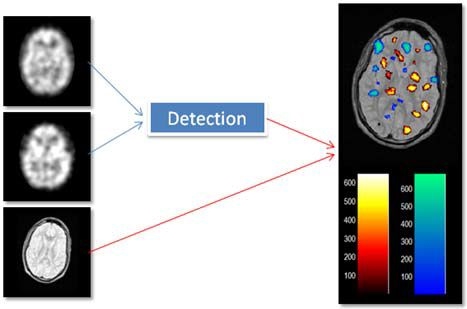
Ictal-interictal SPECT and MR images (left) and analysis result (right).
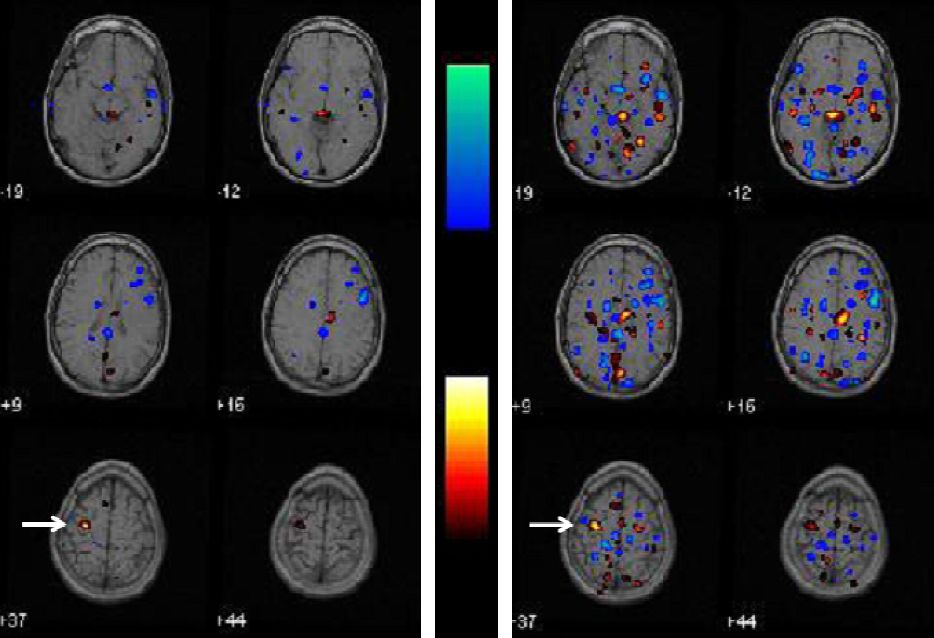
Results for one of the patients Subtraction analysis (right). The EZ in A-contrario detection shows less false activations.
