Direction des Relations Internationales (DRI)
Programme
INRIA "Equipes Associées"
(Dossier
de renouvellement)
|
EQUIPE ASSOCIEE |
Brain Atlas |
|
sélection |
2002 |
|
Renouvellement |
2009 |
|
Equipe-Projet INRIA : Asclepios |
Organisme étranger partenaire : LONI (UCLA) |
|
Centre de recherche INRIA : Sophia-Antipolis
Meditérannée |
Pays : USA |
|
|
Coordinateur français |
Coordinateur étranger |
|
Nom, prénom |
Xavier Pennec |
Paul Thompson |
|
Grade/statut |
DR INRIA |
Professor of Neurology |
|
Organisme d'appartenance |
INRIA, projet ASCLEPIOS |
Lab of Neuro Imaging, UCLA School of Medicine |
|
Adresse postale |
2004 Route des Lucioles, BP 93, |
635 Charles E. Young Drive South, Suite 225E Los Angeles, CA 90095-7332, USA |
|
URL |
||
|
Téléphone |
+33 4 92 38 76 60 |
+1 310 206 2101 |
|
Télécopie |
+33 4 92 38 76 69 |
+1 310 206 5518 |
|
Courriel |
|
Titre de la thématique de collaboration (en français et en anglais) : Brain morphometry / Computational brain anatomy / Brain atlases |
|
Descriptif (environ 10 lignes) : The design of a reference model appropriate for the processing of human brain data is a challenge. Such a model should be able to express the high variability of the shape of structures and the localization of functions within the brain in large populations, take into account notions such as age, sex, pathologies, and be robust to the high dimension-small sample-size (HDSS) problem. The association of the Asclepios and LONI teams is aimed at obtaining a better understanding of the methods to study the structural organization of the brain thanks to the sharing of their experience. Asclepios's strength principally resides in the methodological expertise developed for the robust analysis of medical images and statistics on geometric objects such as surfaces and deformations, while the strength of the LONI lies in a unique international expertise in neuroanatomy and the development and exploitation of large databases of brain images (e.g. key partner of the Human Brain Mapping project). This collaboration allows to factor the methodological expertise on statistical shape analysis and geometric data registration and the clinical databases of a sufficient size needed to to obtain statistically meaningful results (typically several hundreds of images), in order to better understand how the brain varies among subjects and to develop more robust and more accurate brain atlases. Thanks to the combined power of the databases sizes and the precision of the image analysis algorithms, we expect to be able to infer new hypotheses about the underlying structural organization of the brain. |
Understanding and modeling the individual anatomy of human brain and its variability over a population is made difficult by the absence of physical models for comparing different subjects, the complexity of shapes, and the high number of degrees of freedom implied. This raises the need for statistics on objects like curves, surfaces and deformations that do not belong to standard Euclidean spaces. Applications are very important both in neuroscience, to minimize the influence of the anatomical variability in functional group analyses or to discover anatomical differences between populations, and in medical imaging, to better drive the adaptation of generic models of the anatomy (atlas) into patient-specific data.
Anatomy is the science that studies the structure and the relationship in space of different organs and tissues in living systems. Since the 1980ies, an ever growing number of imaging modalities allows observing both the anatomy and the function in vivo and in situ at many spatial scales (from cells to the whole body) and at multiple time scales: milliseconds (e.g. beating heart), years (growth or aging), or even ages (evolution of species). Moreover, the non-invasive aspect allows repeating the observations on multiple subjects. This has a strong impact on the goals of the anatomy which are changing from the description of a representative individual to the description of the structure and organization of organs at the population level. This led in the last 10 to 20 years to the gradual evolution of descriptive atlases into interactive and generative models, allowing the simulation of new observations. For instance, the first brain atlases were based on the detailed description of anatomical specimens [Talairach:67,Talairach:88,Ono:90,Duvernoy:91] or histological preparations revealing the cyto-architecture [Broadman09]. With the growing availability of in vivo images, population-based atlases such as the MNI 305 [Evans:MNI305:1993] and ICBM 152 [Mazziotta:ICBM152:2001] templates were developed and became the basis for the Brain Web MRI simulation engine [Collins:TMI:98]. Similar examples in the orthopedic domain are given by the "bone morphing" method [Fleute:MICCAI:98,Rajamani04:ISBI:04] that simulate the shape of bones.
Building an appropriate reference model to process brain data should allow the expression of the variability of anatomical structures and functions among a large population, take into account the notions of age, sex, the differences induced by pathologies. The method is generally to map some generic (atlas-based) knowledge to patients-specific data through atlas-patient registration. In the case of observations of the same subject, many geometrical and physically based registration methods were proposed to faithfully model and recover the deformations. However, in the case of different subjects, the absence of physical models relating the anatomies leads to a reliance on statistics to learn the geometrical relationship from many observations. The method is to identify anatomically representative geometric features (points, tensors, curves, surfaces, volume transformations), and to model their statistical distribution across the population, for instance via a mean shape and covariance structure analysis after a group-wise matching. In the case of the brain, researchers usually rely on a hierarchy of structural models:
Anatomical or functional landmarks like the AC and PC points [Talairach:88,Bookstein:78];
Curves like crest lines [Subsol98] or sulcal lines [Mangin:TMI:04,LeGoualher:TMI:99,Fillard:NeuroImage:06];
Surfaces like the cortical surface or sulcal ribbons [Thompson:JCAT:97,Andrade:HBM:01,Vaillant:Neuroimage:2007];
Images seen as 3D functions, which lead to voxel-based morphometry (VBM) [Ashburner:VBM:00];
Rigid, multi-affine or diffeomorphic transformations [Trouve:IJCV:98,Miller:ARBE:02,Arsigny:MICCAI:06], leading to Deformation-based or Tensor-based morphometry (TBM) [Chung:NeuroImage01].
One of the question of debate in the community is whether one should compute the variability statistics within the full 3D volume or restricted on the surface of the cortex [VanEssen05,Thompson:Neuroscience:96]. Although this question is of very high interest, we do not plan to investigate it within this project. One of the main reason is that we the new anatomical features which can be extracted from diffusion MRI (see specific section on this subject below) are intrinsically volumetric as they are located in the white matter and the information they provide fade out close to the grey matter and the cortex surface.
The geometric features that are extracted often belong to curved manifolds rather than to Euclidean spaces, which precludes the use of classical linear statistics. For instance, the average of points on a sphere is located inside the sphere and not on its surface. This aspect motivates the research on defining consistent statistical and computing frameworks on Riemannian manifolds. In the case of finite dimensional Riemannian manifolds, we proposed in [Pennec:JMIV:06, Pennec:HDR:06] a set of consistent statistical tools (mean value, covariance matrix, Normal law, Mahalanobis distance) to efficiently work with these features. To remain simple, one of the main features of computing on manifolds is that the mean value has to be defined by the minimization of the expected distance (Fréchet mean). Then, most of the usual algorithms can be rephrased as weighted means, which can be computed iteratively [Pennec:IJCV:05].
An open problem is now to extend this framework to infinite dimensional manifolds such as manifolds of deformations (diffeomorphisms). This is necessary in particular to define properly the mean template and the reference coordinate system in atlas construction. The usual approach consist in alternatively registering all observations to the current template and recomputing the mean template from registered observations. Estimating the reference coordinate system with a known template is generally performed using a gradient descent on a given deformation space. For instance, we defined in [Lepore:MICCAI:07] the as the one minimizing the mean deformation tensor with respect to all observations (see Tensor-based morphometry below). However, when one wants to estimate also the template (the “mean” image), then there are some indications that this simple scheme converges almost surely towards something which is different from the right template [Allassonniere:08]. To avoid this problem, several approaches proposed very recently consider the deformations from the template to the observation as unobserved variables and use a Bayesian approach to estimate the variable of interest (the template) [Allassoniere:JRSS:07, Sabuncu:MICCAI:08, Lepore:MFCA:08]. As this is research problem emerged very recently, there is still a lot of work needed to sort out what has to be done from a theoretical and from a practical point of view.
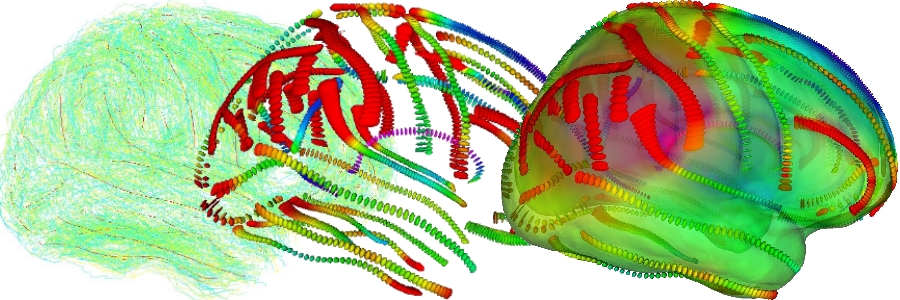
An example of brain variability modeling was provided within this associated team by Pierre Fillard at Asclepios [Fillard:NeuroImage:07], who modeled the variability of the brain from a dataset of anatomical structures (sulcal lines) precisely delineated on the cerebral cortex by the LONI team (see figure below). For each sulcal line, the basic idea to compute the mean lines is to alternatively compute the matches that minimize the distance between the mean curve and the corresponding observation in each subject, and re-estimated the mean curve from the updated matches. For each point of the mean sulcal lines, the variability is then encoded as the covariance matrix of the corresponding anatomical positions in each subject. The sparse field of covariance matrices is then extrapolated to the whole space to obtain a continuous variability field. Statistical tests demonstrated that this model was able to recover the missing information in some places, although it could not fully recover the variability along the sulcal lines. Other results on the correlation between local and distant displacements indicate that the displacement of the symmetric point is correlated and that there are other statistically significant long-distance correlations which were unknown so far [Fillard:WSRPGAAF:07,Fillard:NeuroImage:09].

Figure: From sulcal lines in a population to the brain variability; (left) sulcal lines of eighty subjects in green with the mean sulcal lines in red; (middle) variability measured along the mean sulcal lines (covariance matrix at one sigma); (right) the colour encodes the amount of variability everywhere on the cortex after the extrapolation of the variability tensors onto the whole 3D space, going from blue for the least variable places to red for the most varying ones.
However, this model was known to be limited. In particular, the variability in the direction of the sulcal line itself was minimized on purpose. To overpass this problem and to identify the consequences of this assumption, [Durrleman:MICCAI:07, Durrleman:Media:08] developed a more global approach which finds a diffeomorphism of the whole 3D space that best match the deformed template sulcal lines to the subject ones. A specific representation of lines based on currents was used to get around the point to point correspondence problem between lines. The deformation is belonging to a smooth group of diffeomorphisms provided with the right-invariant metric induced by a reproducible Hilbert space kernel (RKHS) metric on the Lie Algebra of infinitesimal displacement vector fields [Trouve:IJCV:98]. In such a Riemannian framework (see the paragraph statistical computing on manifolds below), each deformation is completely determined by the initial speed vector field, or more concisely by the initial momentum which is non-zero at the template data points only. Thus, the variability itself was measured by the variability of the momentum at each template point. Results showed a good agreement with the previous results at some places, and much more variability at some other places. This demonstrated that one should not rely on one method only to model the brain variability. In order to obtain faithful and unbiased results, we believe that many different sources of information should be investigated (e.g. cortical landmarks like sulcal ribbons and gyri, the surface of internal structures or fibre pathways mapped from Diffusion Tensor Imaging (DTI) for the brain). Likewise, different statistical techniques relying on different assumptions should be used. Individually, these sources of information provide only a partial and biased view of the whole variability. Used jointly, we expect to observe a good agreement in some areas (consensus result), and complementary measures in other areas. This will most probably lead in the near future to new neuroanatomical findings and more robust medical image analysis applications.
To incorporate shape priors in image segmentation or to analyze and classify anatomical differences between groups, one usually define a mean shape and compute the principal modes of the “covariance” matrix via principal component analysis (PCA). From the theoretical point of view, this statistical processing requires some metric properties on the shape space itself. One of the interest of the statistical methods based on currents developed by S. Durrleman is that they can handle not only curves but also surfaces [Durrleman:MICCAI:08, Durrleman:MFCA:08]. Modeling shapes with currents enables us to define an inner product and hence an easy to compute distance between shapes without assuming any point correspondences between discrete structures and without selecting arbitrary features. Since the space of currents is linear, one can compute directly standard statistics on shapes such as mean or PCA.
However, there is a drawback from the computational point if view: the polynomial computational complexity in the number of points in shapes (which was not critical for pairwise registration) becomes a clear bottleneck for groupwise statistics when the template is defined as the “mean” of all the (registered) observations. We proposed in [Durrleman:MICCAI:08] an efficient computational framework that overcomes these limitations by providing a sparse representation of currents at any desired accuracy. Experimental results on 10 meshes of deep brain structures (Caudate, Putamen, Globus Pallidus, Amygdala and Hippocampus for each hemisphere) for 50 subjects (Autism / control) clearly demonstrate the interest of the method: the deformation of a mean obtained from 50 instances is now possible in 5 minutes while it is not previously feasible without high performance computing. This opens the way to the computation of anatomical atlases based on the surface of sub-cortical structures. To avoid introducing a statistical bias in the computation of the template anatomy, we proposed in [Durrleman:MFCA:08] to jointly estimate the template and its deformation in a consistent generative model where observations are considered as noisy version of an unknown template. Compared to the usual backward scheme which estimate a template by pulling back data into a common reference frame, this requires to take into account the Jacobian of the deformations during the atlas computation instead of during its use as a prior. Thus, in addition to better theoretical properties, this also reports some of the cost of the use of statistical shape priors into their construction. The feasibility of the approach was shown by building atlases from 32 sets of the 10 previously described deep brain structures on autistics and controls. However, the statistical power of the method was not sufficient so far to find statistically significant discriminant differences on such a small population.
Thanks to the automatic segmentation methods developed at LONI by Jonathan Morra [Morra:MICCAI:08, Morra:TMI:08], one can now envisage to experiment this framework on a much larger scale. We intend to use in 2009 a database of hippocampuses of 490 subjects scanned twice 1-year apart to investigate the effect of disease effects and/or left/right asymmetry. The 490 people are stratified into Alzheimer' Disease (AD), those at risk for AD (MCI), and healthy controls [Morra:NeuroImage:08a, Morra:NeuroImage:08b]. As the hippocampus is the main system affected by AD, we expect to find strong effects that we could also localize quite precisely. We also plan to add more sub-cortical structures such as the caudate and other deep brain structures during Jon Morra's post-doc in Asclepios in 2010 and 2011 to investigate the correlation between the shape changes in all these structures.
Tensor-based morphometry (TBM) is an increasingly popular method to study differences in brain anatomy statistically In TBM, a non-linear registration algorithm is used to align a set of images to a common space, and a statistical analysis is typically performed on the determinant of the Jacobian matrix on the deformation which encodes for local shrinking or expansions of the underlying virtual material [Ashburner:VBM:00, Chung:NeuroImage01]. In 2005 and 2006, it was proposed to use statistics on the more complete strain tensor instead to better capture differences between populations [Lepore:MICCAI:06] or to consistently learn the shape deformation metric and reuse the result as a regularization penalization for new non-rigid registrations (Riemannian elasticity) [Pennec:MICCAI:05, Pennec:MFCA:06]. As deformation tensors are symmetric positive-definite matrices and do not form a vector space, all computations are performed in the log-Euclidean framework [Arsigny:MICCAI:05]. This mathematical framework was the second main theme developed within this associated team program during the last two years.
Most commonly, one of the control subjects’ images, or a high-resolution single subject MRI atlas, is selected as the reference to which all the other images are mapped [Kochunov:01]. To avoid biases induced by the choosing a single individual as a template, methods for creating an average image using the entire set of controls have also been developed. For instance in [Kochunov:03], a mean template is defined by transforming one of the control images using the average of the displacement fields resulting from its registration to all other controls. A similar approach was also adopted in [Guimond:CVIU:99], where the average was taken with respect to both the deformation and the intensities of the reference images. Groupwise registration is also increasingly common to avoid systematic confounding effects and bias associated with aligning images to a specific individual brain, which can arise when the geometry and intensities of the target image resemble some members of the population more than others. As we said above (see the computing on manifolds section), the template estimation problem raises some important theoretical problems that still need to be sorted out.
In [Lepore:MICCAI:07], we chose to improve the common space itself, by choosing the reference coordinate system that minimizes a natural metric on the deformation from that reference space to the population of control subjects. The natural metric on the deformation is the Riemannian elasticity previously introduced. As the same metric is used to compute the reference space (the mean) and the statistics on the variability (the variance), this method avoids statistical bias. Moreover, it should ease nonlinear registration of new subjects data to a template that is ’closest’ to all subjects’ anatomies. The control brain that is already the closest to ’average’ is found and a gradient descent algorithm is then used to perform the minimization that iteratively deforms this template and obtains the mean shape.
One may wonder that it may be more appropriate to use the mean shape anatomical template derived here in conjunction with registration algorithms whose cost functions are explicitly based on the log-transformed deformation tensors, such as those found for instance in [Brun:WSRPGAAF:07] and [Pennec:MICCAI:05]. To do this, we are currently working on a unified registration and statistical analysis framework in which the regularizer, mean template, and voxel-based statistical analysis are all based on the same log-Euclidean metric (see the 2008 report in this document).
Understanding the architecture of the brain connections and relating it to brain functions is a major goal of neuroimaging. The advances of MR diffusion imaging over the last 10 years have raised the possibility of in vivo investigations into brain circuitry: Diffusion tensor Imaging (DTI) is a unique tool to assess in vivo oriented structures within tissues via the directional measure of water diffusion. First studies in neuroscience focused on the parcellation of the brain according to its connectional architecture in order to infer boundaries between discrete functional regions in the grey matter [Berhens:05]. More recently, the relation between the peripheral white matter and the gray matter was investigated by clustering axonal fiber measurements extracted from DTI [Oishi:NeuroImage:08]. These works suggest that using information from diffusion in conjunction with other data is one of the most promising way to approach brain atlases and inter-subject brain variability. By first understanding the structure of the most important fiber tracts, there is a chance that we could gradually understand more about the organization of the peripheral tracts and finally learn the relationship with the cortical features.
A first step in this process is however to estimate the fiber features from data which are reacher than DTI. Indeed, at the resolution od the current DTI images, one consider that there are fiber crossing in as much as 80% of the brain voxels. Thus, one should rely on methods that allow a higher angular resolution such as High angular resolution diffusion imaging (HARDI). An interesting extension of DTI called tensor distribution function (TDF) has been proposed in [Leow:08] to address fiber crossing and non-Gaussianity in diffusion MR images. By using Gaussian distributions as basis functions, the unknown displacement probability function is expanded with the weights given by the TDF. It can also be viewed as a natural and continuous extension of the tensor mixture model. With the computation of TDF, the water displacement probability function, orientation distribution function (ODF), tensor orientation distribution (TOD), and its corresponding anisotropic properties can all be obtained through simple analytic relations. One of the objective of the BrainAtlas collaboration will be to extend the image processing tools developed for tensors to this model. This new axis of collaboration started in July 2008 during the IPAM summer school in Los Angeles.
As a results of all these tasks, we expect to obtain more complete, more accurate and robust atlases that becomes of routine use in neuroscientific studies.
The laboratory of NeuroImaging (LONI) was established at UCLA in 1987 by Dr. Arthur W. Toga, its current director, to study cerebral metabolism with the goal of understanding the relationship between brain structure and function using image data. Work progressed into three-dimensional reconstruction and visualization. This enabled the study of functional anatomy in the same geometric configuration as that found in the living animal. As these reconstructions became more sophisticated, their application to computational atlases became possible. The construction of brain atlases based on detailed representations of anatomy in a standardized 3D coordinate system is now the major focus. The Laboratory addresses the problem of comparing data across individuals as well as across modalities and increased work in humans began. Work is focused on statistical manipulation of the geometry that makes up the anatomic and functional data sets as well as sophisticated visualizations permitting the communication of the results.
LONI is a key partner in many national and international collaborations, that include universities and academic health centers, and independent research institutions. It acts as the hub of a national neuroimaging resource, directed by Dr. Arthur Toga in the Department of Neurology, which supports over 60 national and international brain imaging collaborations. These collaborations apply novel image analysis approaches to investigate brain structure and function in health and disease. Investigations into brain structure and function require a diverse array of tools to create, analyze, visualize, and interact with models of the brain. The laboratory has a large multi-disciplinary staff have over 15 years of experience in neuroimaging and analyses of brain mapping data. It houses a large super computer, over 50 workstations and a data archival system of over 100 terabytes. The LONI Scientific Visualization Group has a cutting-edge production studio capable of outputting the highest quality video and audio. The wet labs are equipped with state of the art processing equipment for computational neuroanatomy and a suite of optical cameras for functional imaging.
The 4D group at LONI driven by Prof. Paul Thompson is focusing more particularly in developing new mathematical and computational approaches for analyzing human 3D brain image data. Patient populations being studied include large numbers of subjects with Alzheimer's Disease, with mild cognitive impairment, and people at genetic risk for Alzheimer's Disease. Another project studies how HIV/AIDS damages the brain. Other active projects focus on understanding brain changes and drug effects in schizophrenia and several other disorder, and how the brain grows in childhood and in the teenage years. Finally, understanding how our genes (and other factors) affect our brain structure and function can also help us investigate the genetic causes and inherited risks for disease. Part of this work involves constructing population-based brain atlases to encode and represent patterns of anatomic variation, and to detect structural differences in health and disease. These approaches often use some very interesting mathematics as well as high-performance computing techniques. The main mathematical themes are: (1) tensor-based morphometry and nonlinear image registration, (2) Diffusion Tensor Imaging (DTI) and High-Angular Resolution Diffusion Imaging (HARDI), (3) automated structure labeling and pattern recognition, (4) understanding anatomical variability, (5) surface parameterization and matching.
Paul Thompson, Ph.D.: ongoing participation on all the scientific aspects of the collaboration since its inception (group head).
Natasha Lepore Ph.D.: ongoing participation for now three years on tensor-based morphometry and non-linear registration (see publication list).
Alexia Leow, M.D., Ph.D.: collaboration started in 2009 on Tensor Distribution Functions, a potential alternative to current HARDI techniques for the quantification of diffusion images.
Caroline Brun (PhD student): participation for the last two years and for 2009 (until PhD graduation) on non-linear registration (see publication list).
Jonathan Morra (PhD student): participation planned from 2009 to 2010 and later (though a post-doc at Asclepios after the PhD graduation) on learning techniques for understanding anatomical variability.
Agatha Lee (PhD student): collaboration started in 2008 on the analysis of populations of DTI images.
Liang Zhan (PhD student): collaboration started in 2009 on Tensor Distribution Functions, a potential alternative to current HARDI techniques for the quantification of diffusion images.
Paul Thompson is Professor of
Neurology, UCLA since 1994. His educational background includes a
Bachelor of Arts (First Class in Honour Moderations) in Greek &
Latin Languages in 1991 from the University of Oxford (England), a
Master of Arts (Hons.) in Mathematics & Philosophy in 1993 from
the same university, and a Ph.D. from UCLA in neuroscience in 1998 on
Mathematical/Computational
Strategies for Analyzing 3D Human Brain Image Databases.
He is associate editor of Human
Brain Mapping, IEEE Trans. on Medical Imaging, Neuroimage
and member of the editorial board
of Medical Image
Analysis, Cerebral Cortex and
member of the program Committee of numerous international conferences
and workshops.
His research
interests include new strategies for Human Brain Mapping
(mathematical and computer intensive methods, image analysis,
encoding brain variability, building atlases...), the study of
Alzheimer's disease and Schizophrenia though brain imaging, Pediatric
imaging, Neuro-oncology.
Paul Thompson publication
records features 35 refereed journal papers as first-author and
more than 104 as senior or last author, including papers in Nature
Neuroscience, Nature Genetics, IEEE Transactions on Medical Imaging,
IEEE Transactions on Biomedical Engineering, Proceedings of the
National Academy of Sciences, Neuron, Journal of Neuroscience, IEEE
Transactions on Visualization and Computer Graphics, Computer Vision
and Image Understanding, Computer Vision and Pattern Recognition,
NeuroImage, Medical Image Analysis, Elsevier Trends in
Pharmacological Science & Technology, Human Brain Mapping,
Journal of Computer Assisted Tomography, Neuroreport, American
Journal of Psychiatry, Laterality, Journal of Electronic Imaging.
A more complete Vitae including an up-to-date list of collaborations and publications is available here (publications in red are currently submitted).
The irreversible evolution of medical practice toward more quantitative and personalized decision processes for prevention, diagnosis and therapy of diseases is supported by a constantly increasing number of biomedical devices providing in vivo measurements of structures and processes inside the human body (or more generally living systems), at scales varying from the organ to the cellular and even molecular level. Among all these measurements, biomedical images of various forms play a more central role everyday, as well as the exploitation of the genetic information attached to each patient. Facing the need for a more quantitative and personalized medicine based on larger and more complex sets of measurements, the research project-team Asclepios aims at developing: Advanced image analysis tools capable to extract the pertinent information from biomedical images and signals to help diagnosis of diseases; Advanced computational models of the human body and living systems to further interpret this information, predict patient’s evolution and simulate therapy’s effects; Large distributed databases of biomedical images and signals to calibrate and validate previous models. To address all these objectives, the Asclepios research project-team is divided into 4 themes: 1/ Medical Image Analysis; 2/ Biological Image Analysis; 3/ Computational Anatomy; 4/ Computational Physiology.
Despite the advances in the automated analysis of biomedical images, statistical models of the anatomy are still very crude, resulting in poor results in highly deformable regions of the body, or between different subjects. Likewise, very few algorithms actually model the physical or even physiological properties of the human body itself. Coupling biomedical image analysis with statistical, physical and physiological models of the human body, possibly with machine learning procedures, could not only provide a better comprehension of the observed images and signals, but also more efficient tools to detect anomalies. The primary objective of such computational models of the human body is to provide a better understanding of the anatomical variability (Computational Anatomy) and of the major functions of the human body (Computational Physiology). A secondary objective is to provide effective algorithmic tools for their realistic numerical simulations. New methods must be designed to automatically adjust the model parameters to a given person from the available biomedical signals (in particular medical images) and also from prior genetic information. Building such patient-specific models will allow a better prediction of the patient evolution, and a better simulation and evaluation of potential therapies. It remains a challenging goal which requires in particular the resolution of huge inverse problems with massive numbers of measurements and unknowns.
Another important milestone is the development of large databases of subjects and patients including biomedical signals and images as well as genetic information, and the development of specific tools to correlate for instance the shape and evolution of anatomical structures (phenotype) with the genetic information (genotype) and/or pathologies. The construction and exploitation of these databases require the development of specific measurement platforms regrouping cutting edge imaging facilities with easy access provided to internal and external research teams (e.g. the Neurospin platform of CEA or the LONI group at UCLA). The main application areas are in medicine and biology. The development of biomedical image analysis methods combined with computational models of the human body will allow a more profound understanding of the anatomy and physiology of the human body at a much larger scale (both generic and specific) and of the correlation between anatomical or physiological anomalies with the development of pathologies. These models will be helpful to better exploit the huge amount of available biomedical signals (from in vivo molecular and cellular imaging to macroscopic organ imaging) as well as the genetic information potentially available on each patient. A major objective will be to increase the potential for pre-symptomatic diagnosis and early treatment for maximum efficiency.
The objective of Computational Anatomy (CA) is the modeling and analysis of biological variability of the human anatomy. Typical applications cover the simulation of average anatomies and normal variations, the discovery of structural differences between healthy and diseased populations, and the detection and classification of pathologies from structural anomalies. The main current applications are in brain morphometry in close link with neuroscience, although CA is in general not limited to the brain. However, a challenging objective is also to establish new surrogate end-points from the automated analysis of temporal sequences in degenerative central nervous system (CNS) diseases.
Studying the variability of biological shapes is an old problem (cf. the remarkable book "On Shape and Growth" by D'Arcy Thompson). Significant efforts have been made since that time to develop a theory for statistical shape analysis. Despite all these efforts, there is a number of challenging mathematical issues which remain largely unsolved in general. The classical stratification of the problems distinguishes the following 3 levels: 1) extraction from medical images of anatomical reference features (points, curves, surfaces, image intensities, local deformations, fibers...) 2) assignment of a point to point correspondence between these manifolds using a specified class of transformations (e.g. rigid, affine, diffeomorphism); 3) statistical analysis and modeling of anatomical variation from these correspondences. The Asclepiops computational anatomy group focus more particularly on:
Consistent diffeomorphic registration schemes to match the extracted features;
Statistics on the manifolds of these features, specially curves and surfaces;
Linking the resulting anatomical variability results to image analysis algorithms;
Grid-Computing Strategies to exploit large databases.
In this process, the collaboration with world-wide renown teams such as LONI is crucial: 1) to access to large consistent databases of brain images and to carefully edited geometric structures extracted from these images; 2) to validate the results by correlating the statistical findings with the known neuro-anatomy and to disseminate the methods in the neuroscience community.
Xavier Pennec, PhD: principal investigator of the associated team BrainAtlas and responsible of the Computational Anatomy research theme of the Asclepios project-team. Participation since the inception in 2002 to all the aspects of the collaboration, responsible and scientific leader since 2005/2006.
Nicholas Ayache, PhD: participation since the inception to all the aspects of the collaboration.
Stanley Durrleman (PhD student): ongoing participation since 2006 on the study of the anatomical variability of the brain from sulcal lines.
Pierre Fillard, PhD: participation from 2004 to 2007 during his PhD on the study of the anatomical variability of the brain from sulcal lines. Participation is planned to restart if Pierre is recruited as researcher in the Asclepios team (from Sept. 2009 on)
PhD Candidate, to be recruited (in link with the RNTL project Neurolog and an ANR Blanc project BrainVar2 currently in preparation): participation on the statistical analysis of deformations obtained from tensor-based morphometry and on DTI and higher order diffusion analysis methods used as an alternative input to T1 images in population studies.
Xavier Pennec was born on May 2nd, 1970. Dr Pennec is currently a Research Director at the ASCLEPIOS team at INRIA Sophia-Antipolis (France). He holds an Engineering degree from the French Ecole Polytechnique in 1992 and a PhD degree in Computer Science from the same institution in 1996. He was a post-doctoral associate at MIT AI Lab (Mass. USA) in 1997, before joining INRIA in 1998. In 2006, he defended his habilitation at Nice-Sophia Antipolis University. His main research axes are about statistics on geometric data, in particular for medical image analysis, and biomedical image registration. Over the last years, these fields have gradually converged toward computational anatomy, which aims at statistically describing the normal and abnormal shape of organs across populations. X. Pennec co-authored more than 100 peer-reviewed papers in international journals and conferences in these fields. He is associate editor of the Medical Image Analysis Journal (Elsevier); he created and chaired the International Workshop on Mathematical Foundations of Computational Anatomy in 2006 and 2008.
Xavier Pennec was the scientific leader of the INRIA part for the European projects Roboscope and Health-e-Child. Other research grants where he was actively involved include the INRIA ARC BrainVar, the ANR-funded project Neurolog, the ACI AGIR and the associated team Brain Atlas. A more detailed Vitae is available on the web.
Prior to the associated team BrainAtlas, several informal collaborations took place between the two teams, like the evaluation of an histological slice registration method developed at Epidaure thanks to an atlas of a rat brain acquired at the LONI [Ourselin:IVC:2001], or the participation of Epidaure to the writing of the "brain warping" book edited by A. Toga on digital techniques for the analysis of the human brain [Subsol:BrainWarping:1998,Thirion:BrainWarping:1998].
During the first two years of this joint-team project, the PhD thesis of Alain Pitiot [Pitiot:PhD:2003], jointly supervised by N. Ayache and P. Thompson, strengthened the scientific exchanges between the two teams. This thesis was devoted to the segmentation of anatomical structures of the brain for the creation of atlases, and was successfully defended on November 26, 2003. Many common publications were jointly authored by Alain Pitiot and members of both the EPIDAURE and LONI teams between 2002 and 2006.
Since 2004, Vincent Arsigny and Pierre Fillard dedicate part of their PhD to the study of the brain variability using the massive database of images and delineated cerebral structures acquired by P. Thompson at the LONI [Fillard:IPMI:05, Fillard:NeuroImage:06, Fillard:Neuroimage09]. This work was also continued with the PhD of Stanley Durrleman with alternative diffeomorphic registration methods [Durrleman:Media:08].
In 2006, a new dynamic was initiated with the arrival of Natasha Lepore in post-doc and Caroline Brun in PhD at UCLA. Several common research project were started around "Tensor-based Morphometry". Through regular exchanges, this new thematics gave rise to several publications in 2007 and 2008 (see below). In December 2006, X. Pennec defended his habilitation on Statistical computing on manifold for computational anatomy [Pennec:HDR:2006], of which Paul Thompson was reviewers. The framework of the associated team is summarized in this document.
The Summer 2008 was a unique opportunity to foster the active collaborations with C. Brun N. Lepore and and to start new ones involving J. Morra and A. Leow thanks to the very successful IPAM summer school on Mathematics in Brain Imaging organized by P. Thompson in July 2008 (in which X. Pennec, and N. Lepore presented part of the work realized within the associated team framework) and to the International Workshop on Mathematical Foundations of Computational Anatomy organized by X. Pennec in September 2008 in conjunction with MICCAI.
The first objective of 2008 was to finalize the work of P. Fillard and S. Durrleman on the estimation of the anatomical variability of the brain from sulcal lines. The methodology developed to study the joint variability of any pair of cortical positions and its results were finally submitted to Neuroimage and should appear soon [Fillard:NeuroImage:09]. The reason why it was not submitted to Nature in Neuroscience or PNAS is that the results were very difficult to explain and no striking neuroscientific finding was found to put into evidence. Neuroscientific interpretations of the results obtained by S. Durrleman on the sulcal lines were also drawn, with a journal paper as result [Durrleman:Media:08].
The theoretical work on the statistical modeling of curves by S. Durrleman was continued and extended to surfaces. The work done in 2008 is described in details in Section “Modeling the brain variability from the surface of sub-cortical structures” of the scientific goals of the associated team. This work led to one publication at the MFCA workshop (see events below) [Durrleman:MFCA08] and one publication a the MICCAI conference [Durrleman:MICCAI:08] which was awarded the "Young Investigator Award 2008" in the category "Shape and Statistical Analysis". In parallel an important work was performed by J, Morra at LONI to segment sub-cortical brain structures in a large database of almost 500 subjects [Morra:MICCAI:08, Morra:TMI:08, Morra:NeuroImage:08a, Morra:NeuroImage:08b]. We expect that the combination of these two works in 2009 will lead to new advances in the modeling of the brain shape variability.
The work that we initiated last year on tensor-based morphometry was continued on two axes.
Firstly, we studied the influence of the choice of template in tensor-based morphometry [Lepore:ISBI:08]. Using 3D brain MR images from 10 monozygotic twin pairs, we defined a tensor-based distance between each image pair in the study. Relative to this metric, twin pairs were found to be closer to each other on average than random pairings, consistent with evidence that brain structure is under strong genetic control. We also computed the intraclass correlation and associated permutation p-value at each voxel for the determinant of the Jacobian matrix of the transformation. The cumulative distribution function (cdf) of the p-values was found at each voxel for each of the templates and compared to the null distribution. Surprisingly, there was very little difference between CDFs of statistics computed from analyses using different templates. As the brain with least log-Euclidean deformation cost, the mean template defined here avoids the blurring caused by creating a synthetic image from a population, and when selected from a large population, avoids bias by being geometrically centered, in a metric that is sensitive enough to anatomical similarity that it can even detect genetic affinity among anatomies.
Secondly, we continued the development of our fluid registration method that computes the mappings and performs TBM statistics in a consistent way. This was realized by defining a new regularizer that is a fluid extension of the Riemannian elasticity, which assures diffeomorphic transformations. The consistency between TBM registration and statistics should improve the detection power , which is paramount in epidemological studies or drug trials. We applied our method to an MRI dataset from 40 fraternal and identical twins, to reveal voxelwise measures of average volumetric differences in brain structure for subjects with different degrees of genetic resemblance. On the average, the difference in brain structure volumes was found to be less in identical (MZ) than fraternal (DZ) twins [Brun:ISBI:08, Brun:OHBM:08], which is consistent with the degree of genetic similarity. To go one step further, we mapped in [Brun:MICCAI:08] the heritability of brain morphology. Heritability maps were computed from the intraclass correlations and their significance was assessed using voxelwise permutation tests. Lobar volume heritability was also studied using the ACE genetic model. The performance of the Riemannian fluid registration algorithm was compared to a more standard fluid registration algorithm. Results showed that 3D maps from both registration techniques displayed similar heritability patterns throughout the brain. However, by comparing the cumulative distribution functions of the p-values from both methods, the Riemannian algorithm was shown to outperformed the standard fluid registration in terms of statistical power.
Two important scientific events were organized this year in link with the associated team:
P. Thompson organized in July 2008 a very successful IPAM summer school on Mathematics in Brain Imaging. X. Pennec and N. Leporé were invited to give oral presentations in which they detailed in particular the work realized within the associated team framework. S. Durrleman (Asclepios), C. Brun, A. Leow and J. Morra (LONI) also participated to this summer school.
X. Pennec organized for the second time the international workshop on Mathematical Foundations of Computational Anatomy (MFCA'08) in conjunction with the conference MICCAI'2008 in New York on September 6, 2008. This workshop features 4 talks by members of 4D team at LONI including two talks given by A. Leow and one by N. Lepore (see the MFCA'08 proceedings), as well as one talk by S. Durrleman from Ascelpios [Durrleman:MFCA:08, Lepore:MFCA:08].
With the additional help of the NSF-INRIA REUSSI internship program, we hosted the visit of C. Brun (PhD student at UCLA) and L. Lepore (post-doc at UCLA) for three weeks in April/May in the Asclepios team. Right after this stay, they participated to the ISBI conference in Paris with presentations related to the associated team [Brun:ISBI:08, Lepore:ISBI:08] Sophia. They participated as well to the Human Brain Mapping conference in Melbourne, Australia, on June 15-19 [Brun:OHBM:08, Lepore:OHBM:08]. Almost all the participants to this associated team (C. Brun, N. Lepore, A. Leow, J. Morra for LONI and S. Durrleman and X. Pennec for Asclepios) participated to the MICCAI'08 conference in New-York on Sept. 6-10 [Brun:MICCAI:08, Durrleman:MICCAI:08, Morra:MICCAI:08]. This conference was in fact the occasion of the third BrainAtlas meeting in 2008.
The results of the collaboration were also presented by X. Pennec at invited seminars at the Probability and Statistics Lab (LSP) in Toulouse, France on February 19, and at CSAIL, MIT (Biomedical Imaging and Analysis seminar series) on September 12, 2008, as well as at invited plenary talks at the the Journées MAS de la SMAI (french applied and and industrial mathematical society), Rennes, France, August 29 and at Emerging Trends in Visual Computing (ETVC’08), Ecole Polytechnique, November 18th-20th, 2008.
[Brun:WSRPGAAF:07] Caroline Brun, Natasha Lepore,
Xavier Pennec, Yi-Yu Chou, Oscar L. Lopez, Howard J. Aizenstein,
Arthur W. Becker, James T. andToga, and Paul M. Thompson Thompson.
Comparison of Standard and Riemannian
Fluid Registration for Tensor-Based Morphometry in HIV/AIDS.
In Proc. of MICCAI'07 Workshop on
Statistical Registration: Pair-wise and Group-wise Alignment and
Atlas Formation, Brisbane, Australia, 2007.
![]()
[Brun:MICCAI:08] Caroline Brun, Natasha Lepore, Xavier
Pennec, Yi-Yu Chou, Agatha D. Lee, Marina Barysheva, Greig I. de
Zubicaray, Matthew Meredith, Katie McMahon, Margaret J. Wright,
Arthur W. Toga, and Paul M. Thompson. A
tensor-based morphometry study of genetic influences on brain
structure using a new fluid registration method.
In Dimitris Metaxas and Leon Axel, editors, Proc.
MICCAI'08, LNCS 5242, New York, USA, pages
914-921, September 2008. Springer-Verlag.
![]()
[Brun:ISBI:08] Caroline Brun, Natasha Lepore, Xavier
Pennec, Yi-Yu Chou, Agatha D. Lee, Greig I. de Zubicaray, Katie
McMahon, Margaret J. Wright, Marina Barysheva, Arthur W. Toga, and
Paul M. Thompson. A new registration
method based on Log-Euclidean Tensor metrics and its application to
genetic studies. In Proc.
of the 2008 IEEE Int. Symp. on Biomedical Imaging: From Nano to
Macro (ISBI'08), Paris, France, May 14-17,
pages 1115-1118, 2008.
![]()
[Brun:OHBM:08] Brun C, Lepore N, Pennec X, Chou YY, Lee AD, Barysheva M, McMahon K, de Zubicaray GI, Wright M, Toga AW, Thompson PM (2008). Volumetric Differences in Brain Structure in Identical and Fraternal Twins Computed using Riemannian Tensor-Based Morphometry, 13th Annual Meeting of the Organization for Human Brain Mapping (OHBM), Melbourne, Australia, June 15-19, 2008.
[Durrleman:MedIA:08] Stanley Durrleman, Xavier Pennec,
Alain Trouvé, Paul Thompson, and Nicholas
Ayache. Inferring brain variability
from diffeomorphic deformations of currents: an integrative
approach. Medical
Image Analysis, 12/5(12):626-637, 2008.
![]()
![]()
![]()
[Fillard:NeuroImage:09] Pierre Fillard, Xavier Pennec,
Paul M. Thompson, and Nicholas Ayache. Evaluating
Brain Anatomical Correlations via Canonical Correlation Analysis of
Sulcal Lines. NeuroImage,
2009. Note: Accepted for publication.
![]()
[Fillard:WSRPGAAF:07] P. Fillard, X. Pennec, P.M.
Thompson, and N. Ayache. Evaluating
Brain Anatomical Correlations via Canonical Correlation Analysis of
Sulcal Lines. In Proc.
of MICCAI'07 Workshop on Statistical Registration: Pair-wise and
Group-wise Alignment and Atlas Formation,
Brisbane, Australia, 2007.
![]()
[Fillard:NeuroImage:07] Pierre Fillard, Vincent
Arsigny, Xavier Pennec, Kiralee M. Hayashi, Paul M. Thompson, and
Nicholas Ayache. Measuring Brain
Variability by Extrapolating Sparse Tensor Fields Measured on Sulcal
Lines. Neuroimage,
34(2):639-650, January 2007. Note: Also as INRIA Research Report
5887, April 2006. PMID: 17113311.
![]()
![]()
[Fillard:PhD:08] Pierre Fillard. Riemannian
Processing of Tensors for Diffusion MRI and Computational Anatomy of
the Brain. PhD Thesis, University of
Nice-Sophia Antipolis, February 2008.
![]()
[Lepore:ISBI:08] Natasha Lepore, Caroline Brun, Yi-Yu
Chou, Agatha D. Lee, Marina Barysheva, Xavier Pennec, Katie McMahon,
Matthew Meredith, Greig I. de Zubicaray, Margaret J. Wright, Arthur
W. Toga, and Paul M. Thompson. Best
individual template selection from deformation tensor minimization.
In Proc. of the 2008 IEEE Int. Symp. on
Biomedical Imaging: From Nano to Macro (ISBI'08), Paris, France, May
14-17, pages 460-463, 2008.
![]()
[Lepore:MICCAI:07] Natasha Lepore, Caroline Brun,
Xavier Pennec, Yi-Yu Chou, Oscar L. Lopez, Howard J. Aizenstein,
James T. Becker, Arthur W. Toga, and Paul M. Thompson. Mean
Template for Tensor-Based Morphometry using Deformation Tensors.
In Nicholas Ayache, Sébastien Ourselin, and Anthony Maeder,
editors, Proc. Medical Image Computing and
Computer Assisted Intervention (MICCAI'07),
volume 4792 of LNCS,
Brisbane, Australia, pages 826-833, October 2007. Springer. Note:
PMID: 18044645.
![]()
![]()
[Annese:Neuroimages:2004] J. Annese, A. Pitiot, I.D.
Dinov, and A.W. Toga. A
myelo-architectonic method for the structural classification of
cortical areas. NeuroImage,
21(1):15-26, January 2004.
![]()
[Pitiot:PhD:2003] Alain Pitiot. Segmentation
automatique des structures cérébrales s'appuyant sur
des connaissances explicites. Thèse
de sciences, École des mines de Paris, November 2003.
![]()
[Pitiot:JOA:2004] A. MacKenzie-Graham, E.F. Lee, I.D. Dinov, M. Bota, D.W. Shattuck, S. Ruffins, H. Yuan, F. Konstantinidis, A. Pitiot, Y. Ding, G. Hu, R.E. Jacobs, and A.W. Toga. A multimodal, multidimensional atlas of the C57BL/6J mouse brain. Journal of Anatomy, 204(2):93-102, February 2004.
[Pitiot:Neuroimage:2004] A. Pitiot, H. Delingette, P.M.
Thompson, and N. Ayache. Expert
Knowledge Guided Segmentation System for Brain MRI.
NeuroImage,
NeuroImage,
23(supplement 1):S85-S96, 2004. Note: Special Issue: Mathematics in
Brain Imaging.
![]()
[Pitiot:TMI:02] Alain Pitiot, Paul M. Thompson, and
Arthur W. Toga. Adaptive Elastic
Segmentation of Brain MRI via Shape Model Guided Evolutionary
Programming. IEEE
Transactions on Medical Imaging,
21(8):910-923, August 2002.
![]()
[Pitiot:BrainMapping:03] J Annese, A. Pitiot, L.D. Dinov, and A.W. Toga. A myelo-architectonic method for the structural classification of cortical areas. In Human Brain Mapping HBM'03, 2003.
[Pitiot:MICCAI:03] A. Pitiot, H. Delingette, N. Ayache,
and P.M. Thompson. Expert-Knowledge-Guided
Segmentation System for Brain MRI. In
Randy E. Ellis and Terry M. Peters, editors, Medical
Image Computing and Computer-Assisted Intervention MICCAI'03,
volume 2879 of LNCS,
Montreal, pages 644-652, November 2003. Springer Verlag.
![]()
[Pitiot:IPMI:03] A. Pitiot, H. Delingette, A. Toga, and
P. Thompson. Learning Object
Correspondences with the Observed Transport Shape Measure.
In Information Processing in Medical Imaging
IPMI'03, 2003.
![]()
[Pitiot:WBIR:03] A. Pitiot, G. Malandain, E. Bardinet,
and P. Thompson. Piecewise Affine
Registration of Biological Images. In
J.C. Gee, J.B. A. Maintz, and M. W. Vannier, editors, Second
International Workshop on Biomedical Image Registration WBIR'03,
volume 2717 of Lecture Notes in Computer
Science, Philadelphia, PA, USA, pages
91-101, 2003. Springer-Verlag. Note: Also research report INRIA
RR-4866.
![]()
[Pitiot:HBMa:02] J Annese, A. Pitiot, and A. Toga. 3D cortical thickness maps from histological volumes. In Human Brain Mapping HBM'02, 2002.
[Pitiot:HBMc:02] A. Mackenzie-Graham, E. Lee, I. Dinov, A. Pitiot, G. Hu, M. Bota, Y. Ding, L. Capetillo-Cunlife, K. Crawford, B. Truong, and A. Toga. Atlas of the C57BL/6 mouse brain: A multimodal, multidimensional approach. In Human Brain Mapping HBM'02, 2002.
[Pitiot:IJCNN:02] A. Pitiot, A. Toga, N. Ayache, and P.
Thompson. Texture based MRI segmentation
with a two-stage hybrid neural classifier.
In World Congress on Computational
Intelligence / INNS-IEEE International Joint Conference on Neural
Networks WCCI-IJCNN'02, 2002.
![]()
[Pitiot:HBMb:02] S. Ying, J. Alger, A. Pitiot, M. Lee, J. Jen, R. Baloh, D. Perlman, D. Geschwind, M. Jacoboni, J. Mazziotta, and A. Toga. Cerebellar control of oculomotion in patients with neuronal calcium channel mutations: high resolution MRI, genotype-phenotype correlation, and functional anatomical correlation. In Human Brain Mapping HBM'02, 2002.
[Pitiot:Neurology:02] S. Ying, J. Alger, A. Pitiot, D. Rex, J. Jen, R. Baloh, M. Jacoboni, J. Mazziotta, and A. Toga. Stochastic tomography, regional volumetric comparisons, and genotype-anatomical correlation in patients with spinocerebella ataxia type 6 and episodic ataxia type 2. In American Academy of Neurology, 2002.
[Pitiot:HBM:01] J. Annese, A. Pitiot, and A.W. Toga. Complex topological analysis of the human striate cortex (V1). In Human Brain Mapping HBM'01, 2001.
[Pitiot:Neuroscience:01] S. Ying, J. Alger, A. Pitiot, D.E. Rex, J. Jen, R.W. Baloh, M. Jacoboni, J.C. Mazziotta, and A.W. Toga. Stochastic tomography, regional volumetric comparisons, and genotype-anatomical correlation in patients with spinocerebella ataxia type 6 and episodic ataxia type 2. In Society of NeuroImage, 2001.
[Pitiot:RR:2003]
Alain Pitiot, Eric Bardinet, P. Thompson, and G. Malandain.
Automated Piecewise Affine Registration
of Biological Images. Research report
RR-4866, INRIA, July 2003.
![]()
[Arsigny:MICCAI:2003]
Vincent Arsigny, Xavier Pennec, and Nicholas Ayache. Polyrigid
and Polyaffine Transformations: A New Class of Diffeomorphisms for
Locally Rigid or Affine Registration.
In Randy E. Ellis and Terry M. Peters, editors, Proc.
of MICCAI'03, Part II,
volume 2879 of LNCS,
Montreal, pages 829-837, November 2003. Springer Verlag.
![]()
![]()
[Commowick:MICCAI:05]
Olivier Commowick, Radu Stefanescu, Pierre Fillard, Vincent Arsigny,
Nicholas Ayache, Xavier Pennec, and Grégoire Malandain.
Incorporating
Statistical Measures of Anatomical Variability in Atlas-to-Subject
Registration for Conformal Brain Radiotherapy.
In J. Duncan and G. Gerig, editors, Proc.
of MICCAI 2005, Part II,
volume 3750 of LNCS,
Palm Springs, CA, USA, October 26-29, pages 927-934, 2005. Springer
Verlag.
![]()
![]()
![]()
[Arsigny:Brevet:05] Vincent Arsigny, Xavier Pennec, Pierre Fillard, and Nicholas Ayache. Dispositif perfectionné de traitement ou de production d'images de tenseurs. French patent filing number 0503483, April 2005.
[Arsigny:MICCAI:05]
Vincent Arsigny, Pierre Fillard, Xavier Pennec, and Nicholas Ayache.
Fast
and Simple Calculus on Tensors in the Log-Euclidean Framework.
In J. Duncan and G. Gerig, editors, Proc.
of MICCAI 2005, Part I,
volume 3749 of LNCS,
Palm Springs, CA, USA, October 26-29, pages 115-122, 2005. Springer
Verlag.
![]()
![]()
![]()
[Arsigny:RR:05] V.
Arsigny, P. Fillard, X. Pennec, and N. Ayache. Fast
and Simple Computations on Tensors with Log-Euclidean Metrics.
Research Report RR-5584, INRIA, Sophia-Antipolis, France, May 2005.
![]()
![]()
![]()
[Arsigny:SIAMX:06]
Vincent Arsigny, Pierre Fillard, Xavier Pennec, and Nicholas Ayache.
Geometric
Means in a Novel Vector Space Structure on Symmetric
Positive-Definite Matrices.
SIAM
Journal on Matrix Analysis and Applications,
29(1):328-347, 2006.
![]()
![]()
[Arsigny:MICCAI:06] Vincent Arsigny, Olivier Commowick, Xavier Pennec and Nicholas Ayache, A Log-Euclidean Framework for Statistics on Diffeomorphisms. In Proc. of the 9th International Conference on Medical Image Computing and Computer Assisted Intervention (MICCAI'06), Part I, LNCS 4190, p.924—931, Springer 2006.
[Durrleman:MICCAI:07]
Stanley Durrleman, Xavier Pennec, Alain Trouvé, and Nicholas
Ayache. Measuring
Brain Variability via Sulcal Lines Registration: a Diffeomorphic
Approach.
IIn Nicholas Ayache, Sébastien Ourselin, and Anthony Maeder,
editors, Proc.
Medical Image Computing and Computer Assisted Intervention (MICCAI),
volume 4791 of LNCS,
Brisbane, Australia, pages 675-682, October 2007. Springer. Note:
PMID: 18051117.
![]()
[Durrleman:MFCA:08]
Stanley Durrleman, Xavier Pennec, Alain Trouvé, and Nicholas
Ayache. A
Forward Model to Build Unbiased Atlases from Curves and Surfaces.
In X.
Pennec and S. Joshi, editors, Proc. of the International Workshop on
the Mathematical Foundations of Computational Anatomy (MFCA-2008),
September 2008.
![]()
[Durrleman:MICCAI:08]
Stanley Durrleman, Xavier Pennec, Alain Trouvé, and Nicholas
Ayache. Sparse
Approximation of Currents for Statistics on Curves and Surfaces.
In Dimitris
Metaxas, Leon Axel, Gabor Szekely, and Gabor Fichtinger, editors,
Proc.
Medical Image Computing and Computer Assisted Intervention (MICCAI),
Part II,
volume 5242 of LNCS,
New-York, USA, pages 390-398, September 2008. Springer.
![]()
[Fillard:IPMI:05]
Pierre Fillard, Vincent Arsigny, Xavier Pennec, Paul Thompson, and
Nicholas Ayache. Extrapolation
of Sparse Tensor Fields: Application to the Modeling of Brain
Variability.
In Gary Christensen and Milan Sonka, editors, Proc.
of Information Processing in Medical Imaging 2005 (IPMI'05),
volume 3565 of LNCS,
Glenwood springs, Colorado, USA, pages 27-38, July 2005. Springer.
![]()
![]()
![]()
[Fillard:RR:05] P.
Fillard, V. Arsigny, X. Pennec, and N. Ayache. Joint
Estimation and Smoothing of Clinical DT-MRI with a Log-Euclidean
Metric.
Research Report RR-5607, INRIA, Sophia-Antipolis, France, June 2005.
![]()
![]()
![]()
[Fillard:ISBI:06]
P. Fillard, V. Arsigny, X. Pennec, and N. Ayache.
Clinical
DT-MRI estimation, smoothing and fiber tracking with log-Euclidean
metrics.
In Proceedings
of the Third IEEE International Symposium on Biomedical Imaging
(ISBI 2006),
Crystal Gateway Marriott, Arlington, Virginia, USA, pages 786-789,
April 2006.
![]()
[Hua:08] Hua X, Leow AD, Lee S, Klunder AD, Toga AW, Lepore N, Chou YY, Brun C, Chiang MC, Barysheva M, Jack CR, Bernstein MA, Britson PJ, Ward CP, Whitwell JL, Borowski B, Fleisher A, Fox NC, Boyes R, Barnes J, Harvey D, Kornak J, Schuff N, Boreta L, Alexander GE, Weiner MW, Thompson PM, For the ADNI Study (2008). 3D Characterization of Brain Atrophy in Alzheimer’s Disease and Mild Cognitive Impairment using Tensor-based Morphometry, NeuroImage, published online, Feb. 21 2008.
[Leow:08]
Alex D. Leow, Siwei Zhu, Katie McMahon, Greig I. de Zubicaray,
Matthew Meredith, Margaret J. Wright, Paul M. Thompson: The
tensor distribution function. ISBI 2008: 863-866.
![]()
[Lepore:MICCAI:06] Natasha Lepore, Caroline A. Brun, Ming-Chang Chiang, Yi-Yu Chou, Oscar L. Lopez, Howard J. Aizenstein, Arthur W. Toga, James T. Becker, Paul M. Thompson. Multivariate Statistics of the Jacobian Matrices in Tensor Based Morphometry and their application to HIV/AIDS, Proceedings of MICCAI 2006 Part I, LNCS 4190, p191-197, 2006.
[Lepore:MFCA:08]
Natasha Leporé, Caroline Brun, Yi-Yu Chou, Agatha D. Lee,
Marina Barysheva, Greig I. de Zubicaray, Matthew Meredith, Katie L.
McMahon, Margaret J. Wright, Arthur W. Toga, and Paul M. Thompson.
Multi-Atlas
Tensor-Based Morphometry and its Application to a Genetic Study of
92 Twins.
In X.
Pennec and S. Joshi, editors, Proc. of the International Workshop on
the Mathematical Foundations of Computational Anatomy (MFCA-2008),
September 2008.
![]()
[Lepore:TMI:08] Lepore N, Brun CC, Chou YY, Chiang MC, Dutton RA, Hayashi KM, Lu A, Lopez OL, Aizenstein HJ, Toga AW, Becker JT, Thompson PM (2008). Generalized Tensor-Based Morphometry of HIV/AIDS Using Multivariate Statistics on Deformation Tensors, IEEE Transactions on Medical Imaging, 27(1):129-141.
[Lepore:OHBM:08] Lepore N, Vachon P, Lepore F, Chou YY, Voss P, Brun C, Lee AD, Tog AW, Thompson PM (2008). 3D Pattern of Brain Changes in Deaf Subjects Mapped using Tensor-Based Morphometry, 13th Annual Meeting of the Organization for Human Brain Mapping (OHBM), Melbourne, Australia, June 15-19, 2008.
[Morra:MICCAI:08]
Jonathan H. Morra, Zhuowen Tu, Liana G. Apostolova, Amity E. Green,
Arthur W. Toga, Paul M. Thompson: Automatic Subcortical
Segmentation Using a Contextual Model. MICCAI (1) 2008:
194-201.![]()
[Morra:TMI:08] Morra JH, Tu Z, Apostolova LG, Green A, Toga AW, Thompson PM. Comparison of Adaboost and Support Vector Machines for Detecting Alzheimer’s Disease through Automated Hippocampal Segmentation, submitted to IEEE Transactions on Medical Imaging, under revision Feb. 2008.
[Morra:NeuroImage:08a] Morra J, Tu Z, Apostolova LG, Green AE, Avedissian C, Madsen SK, Parikshak N, Hua X, Toga AW, Jack CR, Schuff N, Weiner MW, Thompson PM (2008). Validation of a Fully Automated 3D Hippocampal Segmentation Method Using Subjects with Alzheimer’s Disease, Mild Cognitive Impairment, and Elderly Controls, NeuroImage, 2008 Jul 16.
[Morra:NeuroImage:08b] Morra J, Tu Z, Apostolova LG, Green AE, Avedissian C, Madsen SK, Parikshak N, Toga AW, Jack CR, Schuff N, Weiner MW, Thompson PM (2008). Automated Mapping of Hippocampal Atrophy in 1-Year Repeat MRI Data in 490 Subjects with Alzheimer’s Disease, Mild Cognitive Impairment, and Elderly Controls, NeuroImage, Special Issue on Mathematics in Brain Imaging, ed. Thompson PM, Miller MI, Poldrack R, Nichols T, in press, Oct. 8 2008.
[Pennec:HDR:06]
Xavier Pennec. Statistical
Computing on Manifolds for Computational Anatomy.
Habilitation à diriger des recherches, Université Nice
Sophia-Antipolis, December 2006.
![]()
[Pennec:IJCV:06]
Xavier Pennec, Pierre Fillard, and Nicholas Ayache. A
Riemannian Framework for Tensor Computing.
International
Journal of Computer Vision,
66(1):41-66, January 2006.
![]()
[Pennec:JMIV:06]
Xavier Pennec. Intrinsic
Statistics on Riemannian Manifolds: Basic Tools for Geometric
Measurements.
Journal
of Mathematical Imaging and Vision,
25(1):127-154, July 2006. Note: A preliminary appeared as INRIA
RR-5093, January 2004.
![]()
![]()
[Pennec:MICCAI:05]
Xavier Pennec, Radu Stefanescu, Vincent Arsigny, Pierre Fillard, and
Nicholas Ayache. Riemannian
Elasticity: A statistical regularization framework for non-linear
registration.
In J. Duncan and G. Gerig, editors, Proc.
of MICCAI 2005, Part II,
volume 3750 of LNCS,
Palm Springs, CA, USA, October 26-29, pages 943-950, 2005. Springer
Verlag.
![]()
![]()
![]()
[Pennec:MFCA:06]
Xavier Pennec. Left-Invariant
Riemannian Elasticity: a distance on Shape Diffeomorphisms?.
In X. Pennec and S. Joshi, editors, Proc.
of the International Workshop on the Mathematical Foundations of
Computational Anatomy (MFCA-2006),
pages 1-13, 2006.
![]()
[Pennec:Fillard:BIAMA:08]
Xavier Pennec and Pierre Fillard. Statistical
computing on non-linear spaces for computational anatomy.
In N. Paragios, J. Duncan, and N. Ayache, editors, Biomedical
Image Analysis: Methodologies and Applications.
Springer, 2008. Note: To appear.
![]()
[Pennec:MFCA:08]
Xavier Pennec and Sarang Joshi, editors. Proceedings
of the Second International Workshop on Mathematical Foundations of
Computational Anatomy - Geometrical and Statistical Methods for
Modelling Biological Shape Variability,
New York, USA, September 2008.
![]()
[PennecMFCAProcs:06]
Xavier Pennec and Sarang Joshi, editors. Proceedings
of the First International Workshop on Mathematical Foundations of
Computational Anatomy - Geometrical and Statistical Methods for
Modelling Biological Shape Variability, October 1st, 2006
Copenhagen, Denmark,
2006.
![]()
[Ourselin:IVC:2001]
S. Ourselin, A. Roche, G. Subsol, X. Pennec, and N. Ayache.
Reconstructing
a 3D Structure from Serial Histological Sections.
Image
and Vision Computing,
19(1-2):25-31, January 2001.
![]()
[Stefanescu:MEDIA:04]
R. Stefanescu, X. Pennec, and N. Ayache. Grid
Powered Nonlinear Image Registration with Locally Adaptive
Regularization.
Medical
Image Analysis,
8(3):325-342, September 2004. Note: MICCAI 2003 Special Issue.
![]()
[Subsol:BrainWarping:1998] G. Subsol. Crest Lines for Curve Based Warping. In A. W. Toga, editor, Brain Warping, chapter 13, pages 225-246. Academic Press, 1998.
[Thirion:BrainWarping:1998] J.-P. Thirion. Diffusing Models and Applications. In A. W. Toga, editor, Brain Warping, chapter 9, pages 143-155. Academic Press, 1998
[Thompson:Neuroscience:96] Paul M. Thompson, Craig Schwartz, Robert T. Lin, Aelia A. Khan, and Arthur W. Toga, Three-Dimensional Statistical Analysis of Sulcal Variability in the Human Brain, Journal of Neuroscience 16(13):4261-4274, July1996.
[Talairach:67] J. Talairach and G. Szikla. Atlas d’Anatomie Stereotaxique du Telencephale: Etudes Anatomo-Radiologiques. Masson & Cie, 1967.
[Talairach:88] Talairach and Tournoux, Co-Planar Stereotaxic Atlas of the Human Brain: 3-dimensional Proportional System : an Approach to Cerebral Imaging, Thieme Medical Publishers, New York, 1988.
[Ono:90] M. Ono, S. Kubik, and C. Abernathey. Atlas of the Cerebral Sulci. Thieme, 1990.
[Duvernoy:91] H. Duvernoy. The Human Brain. Springer-Verlag, 1991.
[Broadman:09] Brodman, K. Barth;. Lokalisationslehre der Grosshirnrinde in Ihren Prinzipien Dargestellt auf Grund des Zellenbaues. Leipzig, Germany: 1909.
[Evans:MNI305:1993] Evans, Collins, Mills, Brown, Kelly and Peters, 3D statistical neuroanatomical models from 305 MRI volumes. Proc. IEEE-Nuclear Science Symposium and Medical Imaging Conference, p.1813-1817, 1993.
[Mazziotta:ICBM152:2001] Mazziotta, Toga, Evans, Fox, Lancaster, Zilles, Woods, Paus, Simpson, Pike, Holmes, Collins, Thompson, MacDonald, Iacoboni, Schormann, Amunts, Palomero-Gallagher, Geyer, Parsons, Narr, Kabani, Le Goualher, Boomsma, Cannon, Kawashima, Mazoyer, A probabilistic atlas and reference system for the human brain: International Consortium for Brain Mapping (ICBM). Philos Trans R Soc Lond B Biol Sci vol.356, p.1293—1322, 2001.
[Collins:TMI:98] Collins, Zijdenbos, Kollokian, Sled, Kabani, Holmes, Evans, Design and Construction of a Realistic Digital Brain Phantom, IEEE Transactions on Medical Imaging 17(3):463—468, jun1998.
[Fleute:MICCAI:98] Fleute and Lavallée, Building a Complete Surface Model from Sparse Data Using Statistical Shape Models: Application to Computer Assisted Knee Surgery, in Proc. of Medical Image Computing and Computer-Assisted Interventation (MICCAI'98), LNCS 1496:879—887, Springer, 1998.
[Rajamani04:ISBI:04] Rajamani, Joshi and Styner, Bone model morphing for enhanced surgical visualization, in Proc of IEEE Symp. on Biomedical Imaging: Nano toMacro (ISBI) 2004, Vol. 2, p.1255-1258, apr. 2004
[Bookstein:78] Bookstein, The Measurement of Biological Shape and Shape Change, Lecture Notes in Biomathematics Vol. 24, Springer-Verlag, 1978.
[Subsol98] G. Subsol and J.-Ph. Thirion and N. Ayache, A Scheme for Automatically Building {3D} Morphometric Anatomical Atlases: application to a Skull Atlas. Medical Image Analysis 1(2):37-60, 1998.
[Mangin:TMI:04] J.-F. Mangin, D. Riviere, A. Cachia, E. Duchesnay, Y. Cointepas, D. Papadopoulos-Orfanos, D. L. Collins, A. C. Evans and J. Régis, Object-Based Morphometry of the Cerebral Cortex, IEEE Transactions on Medical Imaging 23(8):968—982, aug. 2004.
[LeGoualher:TMI:99] Le Goualher, E. Procyk, Collins, Venugopal,. Barillot and Evans, Automated extraction and variability analysis of sulcal neuroanatomy, IEEE Transactions on Medical Imaging 18(3):206-217, 1999.
[Thompson:JCAT:97] Thompson, MacDonald, Mega, Holmes, Evans, Toga, Detection and Mapping of Abnormal Brain Structure with a Probabilistic Atlas of Cortical Surfaces, Journal of Computer Assisted Tomography 21(4):567-581, 1997.
[Andrade:HBM:01] Andrade, Kherif, Mangin, J.-F Worsley, Paradis, Simon, Dehaene and Poline, Detection of {fMRI} activation using cortical surface mapping, Human Brain Mapping 12:79-93, 2001.
[Vaillant:Neuroimage:2007] Vaillant, Qiu, Glaunès, J and Miller, Diffeomorphic metric surface mapping in subregion of the superior temporal gyrus NeuroImage 34(3):1149-1159, 2007.
[Ashburner:VBM:00] Ashburner. and Friston, Voxel-Based Morphometry - The Methods, Neuroimage, 11, 805–821 (2000).
[Trouve:IJCV:98] Alain Trouvé, Diffeomorphisms Groups and Pattern Matching in Image Analysis, International Journal of Computer Vision 28(3):213—221, 1998.
[Miller:ARBE:02] Miller, Trouvée and Younes, On the metrics and Euler-Lagrange equations of computational anatomy, Annual Review of Biomedical Engineering, p.375-405, 2003.
[Chung:NeuroImage01] M. K. Chunga, K. J. Worsleyb, T. Paus, C. Cherif, D. L. Collins, J. N. Giedd, J. L. Rapoport and A. C. Evans, A Unified Statistical Approach to Deformation-Based Morphometry, NeuroImage 14(3):595-606, September 2001.
[Berhens:05] T.E.J Behrens* and H Johansen-Berg, Relating connectional architecture to grey matter function using diffusion imaging, Philos Trans R Soc Lond B Biol Sci. 2005 May 29; 360(1457): 903–911.
[VanEssen:05] Van Essen , 2005 D.C. Van Essen, A
population-average, landmark- and surface-based (PALS) atlas of
human cerebral cortex, NeuroImage
28 (3) (2005), pp.
635–662.
![]() .
.
[Sabuncu:MICCAI:08] Mert Rory Sabuncu, Serdar Balci, Polina Golland, Discovering Modes of an Image Population through Mixture Modeling, Proc. Of MICCAI 2008, Part II, LNCS 5242, pp. 381–389, 2008.
[Allasonniere:08] S. Allassonniere, E. Kuhn, A. Trouve.
Bayesian Deformable Models Building Via a Stochastic Approximation
Algorithm: A Convergence Study. Submitted.
![]()
[Allasonniere:JRSS:07] S. Allassonniere, Y. Amit, A.
Trouve. Toward a coherent statistical framework for dense deformable
template estimation, JRSS Vol. 69 Part 1, 2007![]() .
.
[Guimond:CVIU:99] Guimond et al. Average brain models: a convergence study, Computer Vision and Image Understanding 77:192–210, 1999.
[Kochunov:03] Kochunov P et al. An optimized individual target brain in the Talairach coordinate system, Neuroimage 17, (2003) 922–927.
[Kochunov:01] Kochunov P et al., Regional spatial normalization: toward an optimal target, J. Comp. Assist. Tomogr. 25, (2001) 805–816.
[Oishi:NeuroImage:08] Kenichi Oishi et al. Human brain white matter atlas: identification and assignment of common anatomical structures in superficial white matter. NeuroImage. In press. 2008.
© INRIA - mise à jour le 15/08/2008