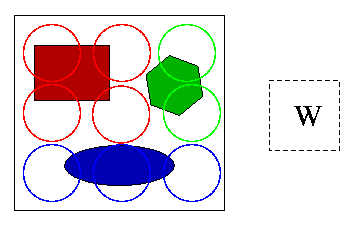
Experimental results (more experiments were conducted in the research
report)
About initialization
We first need to initialize each level set function 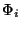 defined in (4). We have
chosen to initialize the
defined in (4). We have
chosen to initialize the 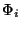 's with circular signed distance
function. We use 2 methods for selecting the initial
's with circular signed distance
function. We use 2 methods for selecting the initial 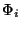 's. The
first one is completely manually handled : we manually select initial
circular level sets.
's. The
first one is completely manually handled : we manually select initial
circular level sets.
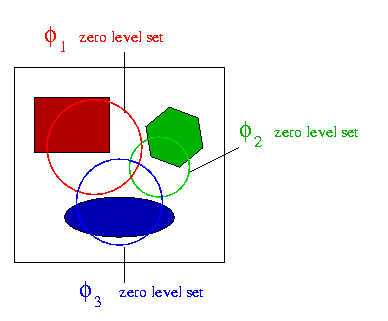
The second one is automatic and we called it "seed
initialization". This method consists of cutting the
data image of u0 into N windows
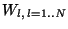 of predefined size. We compute the average ml of u0 on each window
Wl. We then select the index k such that
of predefined size. We compute the average ml of u0 on each window
Wl. We then select the index k such that
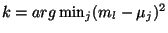 .
And we initialize the corresponding circular signed
distance function
.
And we initialize the corresponding circular signed
distance function 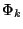 on each Wl. Windows are not overlapping and
each of them is supporting one and only one function
on each Wl. Windows are not overlapping and
each of them is supporting one and only one function 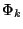 ,
therefore we
avoid overlapping of initial
,
therefore we
avoid overlapping of initial 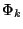 's. The size of the windows is
related to the smallest details we expect to detect. The major avantages of this
simple initialization method are : it is automatic (only the size of
the windows has to be fixed), it accelerates the speed of convergence
(the smaller the windows, the faster the convergence), and it is less
sensitive to noise (in the sense that we compute the average ml of u0over each window before selecting the function
's. The size of the windows is
related to the smallest details we expect to detect. The major avantages of this
simple initialization method are : it is automatic (only the size of
the windows has to be fixed), it accelerates the speed of convergence
(the smaller the windows, the faster the convergence), and it is less
sensitive to noise (in the sense that we compute the average ml of u0over each window before selecting the function 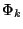 whose mean
whose mean 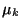 is the closest one to ml).
is the closest one to ml).