|
 |
| Fig. 1: Point light stimuli showing different actions (Demo by R. Blake) | Fig. 2: In spite of strong blurring the action in this video can be recognized (Demo: L. Davis & A. Bobick) |
 |
 |
| Fig. 1: Point light stimuli showing different actions (Demo by R. Blake) | Fig. 2: In spite of strong blurring the action in this video can be recognized (Demo: L. Davis & A. Bobick) |
Input Motion Video Sample |
 |
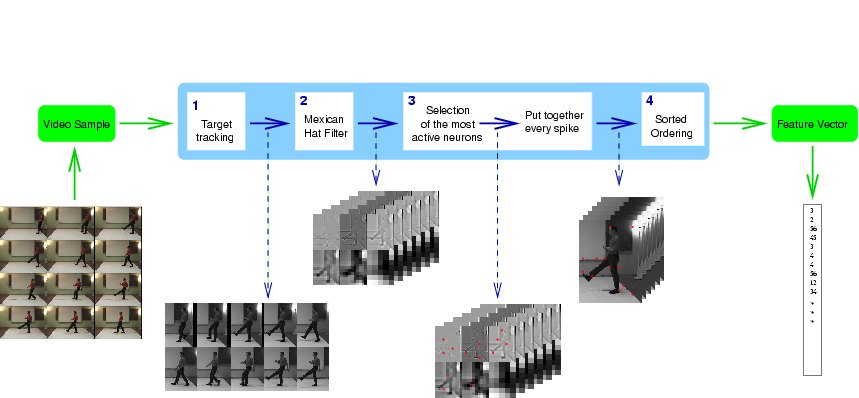
| 10 Spikes | 20 Spikes | 30 Spikes | 40 Spikes |
 |
 |
 |
 |
| 10 Spikes | 20 Spikes | 30 Spikes | 40 Spikes |
 |
 |
 |
 |