Integrated Models of Paediatric Heart Diseases
I- Knowledge-Based Models of Paediatric Heart Diseases
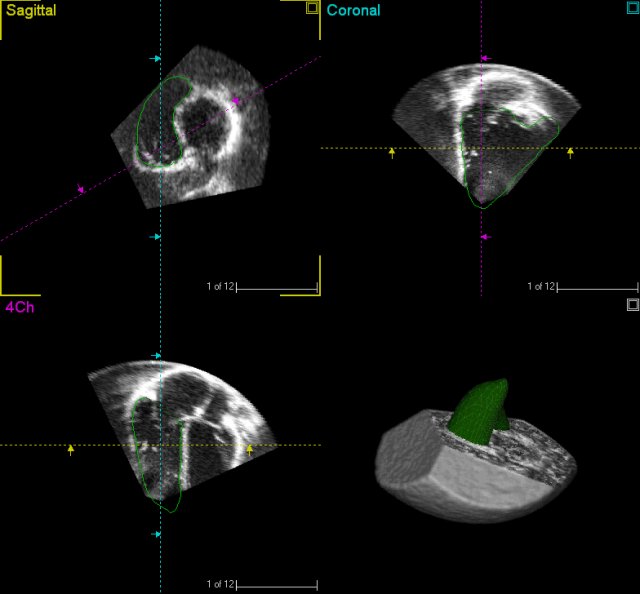
ultrasonography images. (Image from Gaslini)
Abstract. During the diagnostic process, cardiologists and clinicians perform several measurements to identify the pathologies and to assess their severity. One of the main purposes of Health-e-Child is to gather the knowledge and the experience of the cardiologists involved in the project and to design decision-support models that can help the diagnostic of paediatric heart pathologies. In this section we present the underlying principles of a preliminary knowledge-based model that is currently in development. It mainly relies on a set of representative measurements and on a model that formalises the clinical methods used during patients evaluation to automatically identify and classify heart pathologies.
II- Disease-Based Models of Cardiac Electromechanics

Abstract. In the last decades the computational modelling of the human body, and in particular of the heart, has attracted increasing amounts of interest (Ayache, 2004) as it has benefited from progresses in biology, physics and computer science. The understanding of the heart functions, from the heart anatomy to the electrical activity to the biomechanical phenomena, is crucial for the development of advanced treatments of heart diseases. We describe here an in-silico 3-D model for the simulation of the heart function in healthy and ill children. Based on the approach proposed by (Sermesant et al., 2006), this model includes knowledge coming from various disciplines (anatomy, electrophysiology and biomechanics) in a framework that allows direct evaluation with respect to actual clinical measurements.
The model is first adjusted to the physiology of the normal heart in order to calibrate the parameters. Then, preliminary simulations of the paediatric cardiac diseases considered in Health-e-Child are performed by modifying the appropriate macroscopic parameters. Although basic, these simulations provide promising results, the clinical parameters vary accordingly with respect to the simulated pathology.
- Electromechanical model of the heart
- Simulation of the normal heart
- Simulation of right-ventricle overload
- Simulation of dilated cardiomyopathy
- Simulation of hypertrophic cardiomyopathy
- Conclusion
III- Patient-Specific Anatomical Models of the Heart for Decision Support System
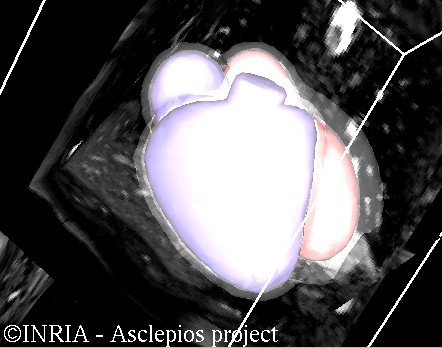
Abstract. In the previous sections two different methods have been described for the assessment and the simulation of paediatric heart diseases. On one hand, there are the models that rely exclusively on parameters representing the patient's physiology; and on the other hand, there are the models where only general considerations are used. Albeit efficient, these two methods do not take advantage of all the available information. The understanding of paediatric cardiac pathologies would greatly benefit from the combination of the two approaches.
An effort must then be done to adapt to the patient physiology the general framework described in the second section of this website. A common approach consists in using 3-D imaging and segmentation techniques to directly create a 3-D representation of the patient's anatomy. However, these methods are often limited by the quality of the underlying image. Another strategy would be to build a geometrical model, as in the disease-based models, but by adapting the synthetic geometry to clinical parameters measured during clinical evaluation. Such an approach would allow us to get a good approximation of the patient's heart anatomy without the requirement of any high-resolution 3-D image.
- Current method: From MRI segmentation to 3-D anatomical model of the heart
- Perspective: From 1-D clinical parameters to 3-D anatomical model of the heart
IV- Personalised Simulation of Myocardium Electromechanics and Pulmonary Valve Replacement Surgery in Repaired Tetralogy of Fallot: a Case Study
Abstract. So far, we have presented on the one hand an electromechanical model of the heart, which is able to perform generic simulation of congenital heart diseases, and on the other hand we described methods to segment the heart from medical images. We investigate in this section how we can use both tools to perform personalised simulations of the heart, in a Health-e-Child patient, and simulate therapies.
We focus on repaired Tetralogy of Fallot. For these patients, pulmonary valve replacement (PVR) is an effective treatment. The therapy consists in replacing the pulmonary valves, surgically or through minimally invasive approaches. The surgeon can also directly remodel the right ventricle to remove any impaired tissue. However, the postoperative outcomes of these techniques mostly depend on patient patho-physiology. We propose in this section an integrated workflow to simulate and predict the effects of PVR upon the cardiac function of a patient. We first personalise an active, anisotropic, visco-elastic electromechanical model of the heart. The two different interventions are then simulated. PPVR is achieved by disabling the regurgitations. Volume reduction surgery is simulated using a soft-tissue intervention platform. Tested on a young patient, the framework showed promising results. Preoperative simulations were successfully personalised (reference RV ejection fraction (EF): 41%, simulated EF: 40%). Postoperative simulations showed that, on this specific patient, the best improvement in cardiac function was achieved by PVR with surgery (EF after PPVR: 40%, after PVR with surgery: 51%).
- Introduction: Medical Background and Challenges
- Creating a Personalised Anatomical Model of the Heart
- Personalising an Electromechanical Model to Simulate Patient Cardiac Function
- Simulating Pulmonary Valve Replacement Therapies
- Conclusions
NEW Automatic Estimation of 3D Myocardium Strain from Anatomical Cardiac Images NEW
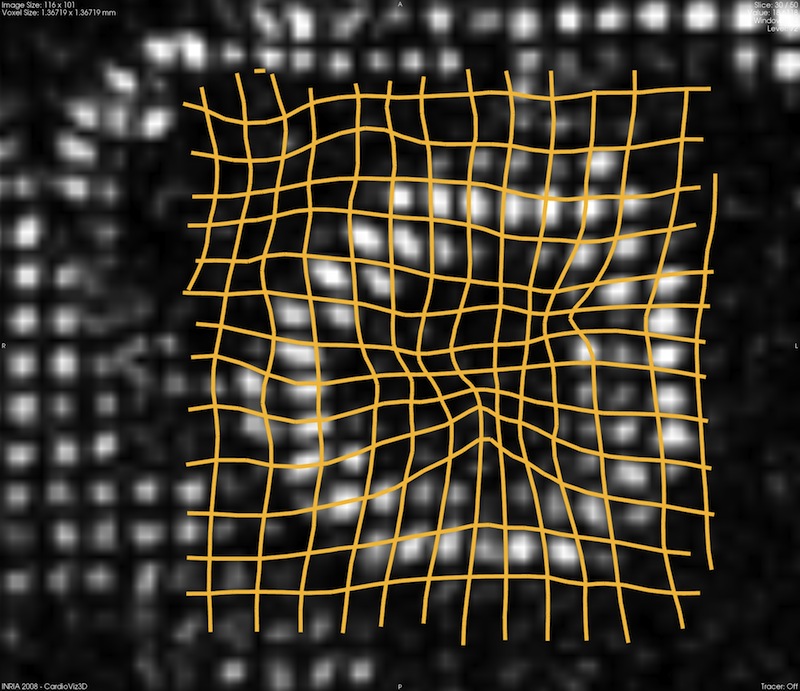
(Data courtesy King's College London, St Thomas Hospital,
Division of Imaging Sciences)
Abstract. The heart is a moving organ and evaluating its intrinsic deformation is crucial for a comprehensive evaluation of its condition. Imaging modalities have been developed to measure the cardiac deformation but when these technologies are not available, little can be provided to the clinicians to assist his diagnostic. Unfortunately, this is the typical situation in congenital heart diseases like tetralogy of Fallot, where only echocardiography or standard cMRI are available so far. In Health-e-Child we tackle the objective of estimating the cardiac deformation from dynamic anatomical images like standard cine MRI. To that end, we constrain a demons-based non-linear image registration algorithm to provide incompressible deformations, a major feature of the myocardium, to cope with the scarce textures and the low image quality. This leads us to a well-posed framework with new insights on demons regularisation. This section describes the overall methodology and an application on a real case. Reported results are published in (Mansi et al., 2010)
- Introduction and Method: iLogDemons, a Consistent Incompressible Image Registration Algorithm
- Experiments on Synthetic Data
- Experiment on a Real Case
References
- Ayache, N., Ed., 2004. Computational Models for the Human Body, ser. Handbook of Numerical Analysis, P. Ciarlet, Ed. Amsterdam, The Netherlands: Elsevier.
-
Sermesant, M., Delingette, H., Ayache, N., 2006. An Electromechanical
Model of the Heart for Image Analysis and Simulation. IEEE
Trans. Med. Imag., vol. 25, no. 5, pp. 612-625.

- T. Mansi, X. Pennec, M. Sermesant, H. Delingette, and N. Ayache. LogDemons Revisited: Consistent Regularisation and Incompressibility Constraint for Soft Tissue Tracking in Medical Images. In Medical Image Computing and Computer Assisted Intervention (MICCAI), Lecture Notes in Computer Science. Springer, 2010. In press.